| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,745,971 – 1,746,063 |
| Length | 92 |
| Max. P | 0.772590 |

| Location | 1,745,971 – 1,746,063 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 73.79 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -17.00 |
| Energy contribution | -17.12 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
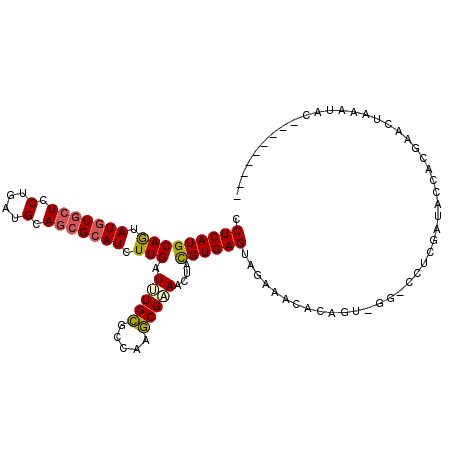
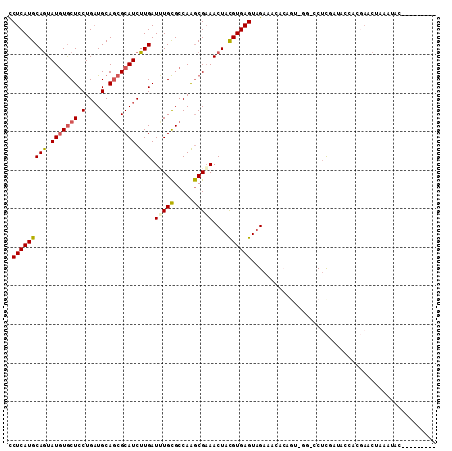
>2L_DroMel_CAF1 1745971 92 + 22407834 CCUCAUGCAGUAUGUGCUCCUGAUGCAGCGCAUCUUGAUUUGCGCCAAGCGAAACUACGUGAGUAGAAA----------CCUCCAUAUCACGAACUAAAUUC--------- .((((((.((((((((((.(....).))))))).....(((((.....)))))))).))))))......----------.......................--------- ( -21.30) >DroSec_CAF1 2629 102 + 1 CCUCAUGCAGUAUGUGCUCCUGAUGCAGCGCAUCUUGAUCUGCGCCAAGCGAAACUACGUGAGUAGAAACACAGUCGAACCUCGAUACCACGAACUGAGUAC--------- .......((((.((((...(((.(((.(((((........)))))...)))...((((....)))).....)))(((.....)))...)))).)))).....--------- ( -27.00) >DroSim_CAF1 9566 102 + 1 CCUCAUGCAGUAUGUGCUCCUGAUGCAGCGCAUCUUGAUCUGCGCCAAGCGAAACUACGUGAGUAGAAACACAGACGGACCUCGAUACCACGAACCAAGUAC--------- .....(((.((.((((.......(((.(((((........)))))...)))...((((....))))...)))).))((...(((......))).))..))).--------- ( -24.10) >DroEre_CAF1 2649 111 + 1 UCUCAUGCAGUAUGUGCUCCUGAUGCAGCGCAUCUUGAUUUGCGCCAAGCGAAACUACGUGAGUAGAAAUACAGUGGGGACUCAAUACCACGAACUAGAUAUCAUGCGAAU ((.(((((((.(((((((.(....).))))))).))).(((((.....))))).((((....)))).......((((..........))))...........)))).)).. ( -29.70) >DroWil_CAF1 18866 74 + 1 CCUCAUGCAAUAUUUACUCCUGCUGCAACGCAUCUUGAUGUGUGCCAGACGUAACUAUGUGAGUUGACA----------ACUUA--------------------------- .(((((...(((.((((..(((..(((.(((((....)))))))))))..)))).))))))))......----------.....--------------------------- ( -16.70) >DroYak_CAF1 9712 103 + 1 CCUCAUGCAGUAUGUGCUCCUGAUGCAGCGCAUCUUGAUUUGCGCCAAGCGAAACUACGUGAGUAGAAACACAGUGGGGUCAUGAU------AACUAAUUAUCAUGCAA-- ((((((.(((.(((((((.(....).))))))).))).(((((.....))))).((((....)))).......)))))).((((((------((....))))))))...-- ( -33.70) >consensus CCUCAUGCAGUAUGUGCUCCUGAUGCAGCGCAUCUUGAUUUGCGCCAAGCGAAACUACGUGAGUAGAAACACAGU_GG_CCUCGAUACCACGAACUAAAUAC_________ .(((((((((.(((((((.(....).))))))).))).(((((.....)))))....))))))................................................ (-17.00 = -17.12 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:46 2006