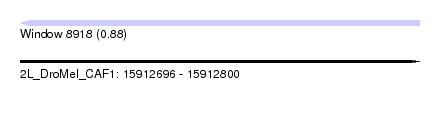
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,912,696 – 15,912,800 |
| Length | 104 |
| Max. P | 0.883119 |

| Location | 15,912,696 – 15,912,800 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -19.83 |
| Energy contribution | -20.75 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15912696 104 - 22407834 CACCACUUCUAGAUCCUAAUUGGCACCGCUACCAACCUAAAACCCACAAGCAGCCACCACGAGGCCUCAAUUGCACUUUGUGGUUGGUUGGCUUGGCGGUGCGG ......................(((((((((((((((......((((((((((((.......)))......)))...))))))..))))))..))))))))).. ( -36.30) >DroSec_CAF1 88962 100 - 1 CACCACUUCUGGAUCCUAAUUGGCACCGCCACCGACCUAAAACCCACAACCAGCCACCACGAGGCCUCAAUUGCACUUUGUGGU----UGGCUUGGUGGUGCGG ..(((....)))..........(((((((((....................(((((((((((((...........)))))))).----)))))))))))))).. ( -33.75) >DroSim_CAF1 90163 100 - 1 CACCACUUCUGGAUCCUAAUUGGCACCGCCACCGACCUAAAACCCACAAGCAGCCACCACGAGGCCUCAAUUGCACUUUGUGGU----UGGCUUGGCGGUGCGG ..(((....)))..........(((((((((....................(((((((((((((...........)))))))).----)))))))))))))).. ( -35.65) >DroEre_CAF1 76980 98 - 1 CACCACUUCUACAUUACAAUACUUACCGCCACCCACC-AAAACCCCCGAGCAGCCACCACGAGGCCUCAAUUGCACUUUGUGGU----UGG-UUGGCGGUGCGG .....................(.((((((((((.(((-.........(((..(((.......))))))....((.....)))))----.))-.)))))))).). ( -26.70) >DroYak_CAF1 68717 100 - 1 CCCCACUUUUAGAUCCUAAUUGGCACCGCCGCCGACCUAAAACCUCCGAUCAGUCACCACGAGACCUCAAUUGCACUUUGUAGU----UGGCUUGGCAGUGCGG ..((.(..((((...))))..)((((.(((((((((................(((.......)))......(((.....)))))----))))..))).)))))) ( -23.60) >DroAna_CAF1 15931 91 - 1 CACC-CUUUACAUUUCUAAUUGGCACCACG--------AAAGCCACGAAGAUGCCGCCACAACGCCUCAAUUACACUUUGUGGU----UUUAAAGGCGGCGUAG ....-...(((...(((...((((......--------...))))...))).((((((..((.(((.(((.......))).)))----.))...))))))))). ( -24.70) >consensus CACCACUUCUAGAUCCUAAUUGGCACCGCCACCGACCUAAAACCCACAAGCAGCCACCACGAGGCCUCAAUUGCACUUUGUGGU____UGGCUUGGCGGUGCGG ......................(((((((((....................(((((((((((((...........))))))))).....))))))))))))).. (-19.83 = -20.75 + 0.92)


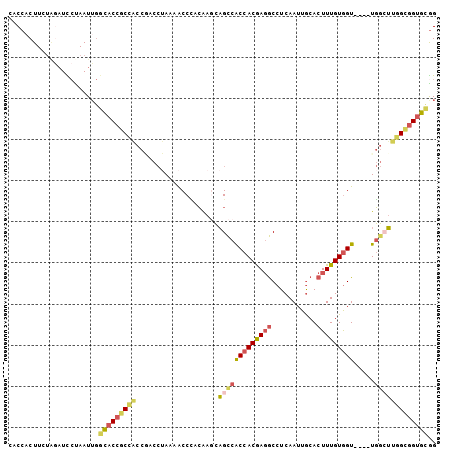
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:43 2006