| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,744,897 – 1,744,989 |
| Length | 92 |
| Max. P | 0.887070 |

| Location | 1,744,897 – 1,744,989 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 73.05 |
| Mean single sequence MFE | -31.13 |
| Consensus MFE | -20.58 |
| Energy contribution | -21.72 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.887070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
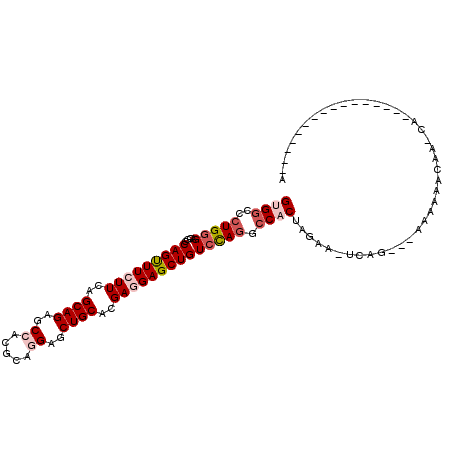
>2L_DroMel_CAF1 1744897 92 - 22407834 GUGGCCCUGCGACGCCAGUUUCUUCAGCAGAGCCACGCAGGAGCUGCACGAGGAGCUGUCCAGGCCACUGGAA-UCCG---AACAAACGAGCAGAA-------------- ......((((..((.(((((((((..((((..((.....))..))))..)))))))))(((((....))))).-....---......)).))))..-------------- ( -33.60) >DroVir_CAF1 1535 93 - 1 GUGUCCCUGGGAAGCCAGUUUCUUCAGCAGCGCCACGCAGGAGCUGCACGAGGAGCUGUCCAGUCCUCUAUAAAAAAUAAGUGGAGUUAAACA----------------- ......(((((....(((((((((..(((((.((.....)).)))))..))))))))))))))..((((((.........)))))).......----------------- ( -31.00) >DroPse_CAF1 1987 87 - 1 GUGGCCCUGGGAGGCCAGUUUUUUCAGCAGCGCCACACAAGAACUGCACGACGAGCUGUCUAGACCACUAAAAAUGAG---AGAAAACA--------------------A .(((((......)))))((((((((.((((..(.......)..))))..(((.....))).................)---))))))).--------------------. ( -22.90) >DroSim_CAF1 8514 106 - 1 GUGGCCCUGGGACGCCAGUUUCUUCAGCAGAGCCACGCAGGAGCUGCACGAGGAGCUGUCCAGGCCACUGGAA-UCCG---AACAAGCGAGCAGAACGACCCGGUCAUUA ((((((..(((.((((((((((((..((((..((.....))..))))..)))))))))(((((....))))).-....---.....))).(.....)..))))))))).. ( -39.50) >DroYak_CAF1 8582 106 - 1 GUGGCCCUGGGACGCCAGCUUCUUCAGCAGAGCCACGCAGGAGCUGCACGAGGAGCUGUCCAAGCCUCUGGAA-UCCG---AAAAAACAACCAGAACGACCCGGCAAUUA ...(((..(((.((.(((((((((..((((..((.....))..))))..)))))))))........(((((..-....---.........))))).)).))))))..... ( -36.86) >DroPer_CAF1 2000 87 - 1 GUGGCCCUGGGAGGCCAGUUUUUUCAGCAGCGCCACACAAGAACUGCACGACGAGCUGUCUAGACCACUAAAAAUGAG---AGAAAACA--------------------A .(((((......)))))((((((((.((((..(.......)..))))..(((.....))).................)---))))))).--------------------. ( -22.90) >consensus GUGGCCCUGGGACGCCAGUUUCUUCAGCAGAGCCACGCAGGAGCUGCACGAGGAGCUGUCCAGGCCACUAGAA_UCAG___AAAAAACAA_CA________________A ((((..(((((....(((((((((..((((..((.....))..))))..)))))))))))))).)))).......................................... (-20.58 = -21.72 + 1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:45 2006