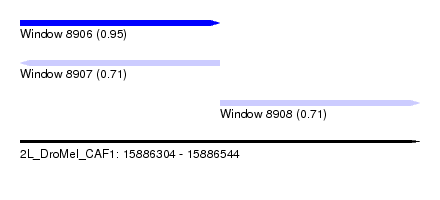
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,886,304 – 15,886,544 |
| Length | 240 |
| Max. P | 0.945887 |

| Location | 15,886,304 – 15,886,424 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.83 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -24.82 |
| Energy contribution | -26.46 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
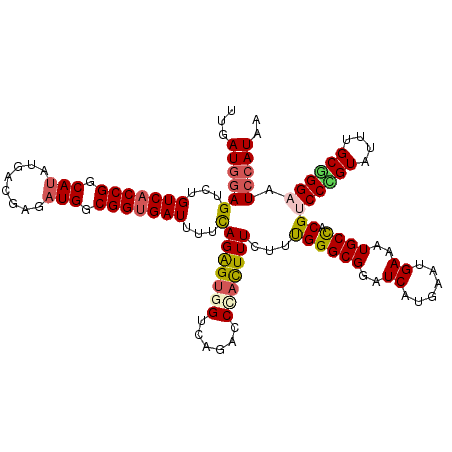
>2L_DroMel_CAF1 15886304 120 + 22407834 UUUUUCUAUGAUUAUUGUUUUUUCCGGUGGUGCCAAACUACGACGAUUCAACCUUUGAACGUUAAUUAAUACGGCCUUAUUGACGUACUUUUGGACGCCAUCGGCCUCCACUUUCAGUAU ............(((((......((((((((((((((.(((((((.((((.....)))))))...((((((......))))))))))..))))).)))))))))..........))))). ( -29.09) >DroSec_CAF1 59746 120 + 1 UUUUUCUAUGAAAAGUGAUUUUUCCGGUGGUGCCAAACUACGUCGUUUCAACCUUUGAACAUUAAUUAAUACGGCUUUGUUGACGUACUUUUGGACGCCAUCGGCCUCCACUUUUAGUAU ......(((.(((((((......((((((((((((((.(((((((...(((((.((((.......))))...))..))).)))))))..))))).)))))))))....))))))).))). ( -35.60) >DroSim_CAF1 59942 120 + 1 UUUUUCUAUGAAAAGUGAUUUUUCCGGUGGUGCCAAACUACGUCGUUUCAACCUUUGAACAUCAAUUAAUACGGCUUUGUUGACGUACUUUUGGACGCCAUCGGCCUCCACUUUUAGUAU ......(((.(((((((......((((((((((((((.(((((((...(((((.((((.......))))...))..))).)))))))..))))).)))))))))....))))))).))). ( -35.60) >DroEre_CAF1 49306 120 + 1 UUCUUCUAUGACAAGUGUCUUCUCCGGCGGCGCCAGACUUCGUCGCUUCCGCCUUUGAACGGUAAUUAAUACGGCUUUGUUGACGUACUUUGGGACGCCUUCGGCCUCCACUUUCAGUAU ........(((.(((((......((((.(((((((((...(((((.....(((.((((.......))))...))).....)))))...)))))..)))).))))....)))))))).... ( -30.50) >DroYak_CAF1 37664 120 + 1 UUCUUCCACGAUAAGUGAUUUUUCCGGCGGUGCCAGACUACGUCGCGUCAACCUUUGAACGUUAAUUAAUACGGCUUUGUUGACGUACUUUGUUACGCCAUCGGCCUCCACUUUCAGUAU .......((...(((((......(((..((((...(((...((.((((((((.......(((........))).....))))))))))...))).))))..)))....)))))...)).. ( -29.20) >consensus UUUUUCUAUGAAAAGUGAUUUUUCCGGUGGUGCCAAACUACGUCGCUUCAACCUUUGAACGUUAAUUAAUACGGCUUUGUUGACGUACUUUUGGACGCCAUCGGCCUCCACUUUCAGUAU ......(((...(((((......((((((((((((((.(((((((......((.((((.......))))...))......)))))))..))))).)))))))))....)))))...))). (-24.82 = -26.46 + 1.64)

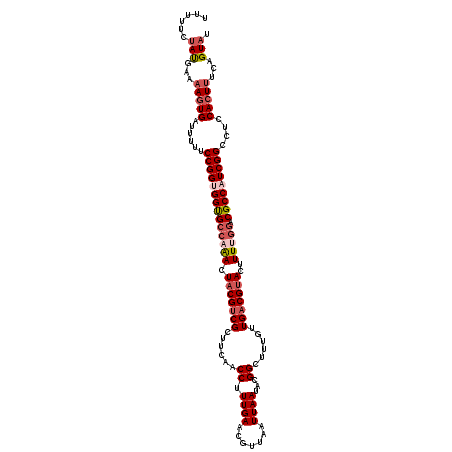
| Location | 15,886,304 – 15,886,424 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.83 |
| Mean single sequence MFE | -30.84 |
| Consensus MFE | -23.21 |
| Energy contribution | -24.21 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15886304 120 - 22407834 AUACUGAAAGUGGAGGCCGAUGGCGUCCAAAAGUACGUCAAUAAGGCCGUAUUAAUUAACGUUCAAAGGUUGAAUCGUCGUAGUUUGGCACCACCGGAAAAAACAAUAAUCAUAGAAAAA ...(((...((((.((((..((((((.(....).))))))....))))((...((((((((((((.....)))).)))..)))))..)).)))))))....................... ( -27.40) >DroSec_CAF1 59746 120 - 1 AUACUAAAAGUGGAGGCCGAUGGCGUCCAAAAGUACGUCAACAAAGCCGUAUUAAUUAAUGUUCAAAGGUUGAAACGACGUAGUUUGGCACCACCGGAAAAAUCACUUUUCAUAGAAAAA ...((((((((((...(((.(((.(.(((((..((((((.....((((...................)))).....)))))).)))))).))).))).....)))))))...)))..... ( -32.31) >DroSim_CAF1 59942 120 - 1 AUACUAAAAGUGGAGGCCGAUGGCGUCCAAAAGUACGUCAACAAAGCCGUAUUAAUUGAUGUUCAAAGGUUGAAACGACGUAGUUUGGCACCACCGGAAAAAUCACUUUUCAUAGAAAAA ...((((((((((...(((.(((.(.(((((..((((((.....((((...................)))).....)))))).)))))).))).))).....)))))))...)))..... ( -32.31) >DroEre_CAF1 49306 120 - 1 AUACUGAAAGUGGAGGCCGAAGGCGUCCCAAAGUACGUCAACAAAGCCGUAUUAAUUACCGUUCAAAGGCGGAAGCGACGAAGUCUGGCGCCGCCGGAGAAGACACUUGUCAUAGAAGAA .((((....(.(((.(((...))).))))..))))((((.....(((.(((.....))).))).....((....))))))...((((((...))))))...(((....)))......... ( -32.90) >DroYak_CAF1 37664 120 - 1 AUACUGAAAGUGGAGGCCGAUGGCGUAACAAAGUACGUCAACAAAGCCGUAUUAAUUAACGUUCAAAGGUUGACGCGACGUAGUCUGGCACCGCCGGAAAAAUCACUUAUCGUGGAAGAA ...(((...(((..((((..((((((.....((((((.(......).)))))).....))).)))..))))..)))....)))((((((...))))))....((((.....))))..... ( -29.30) >consensus AUACUGAAAGUGGAGGCCGAUGGCGUCCAAAAGUACGUCAACAAAGCCGUAUUAAUUAACGUUCAAAGGUUGAAACGACGUAGUUUGGCACCACCGGAAAAAUCACUUAUCAUAGAAAAA ...(((.(((((....(((.(((.(((..(((.((((((.....((((...................)))).....)))))).)))))).))).)))......)))))....)))..... (-23.21 = -24.21 + 1.00)

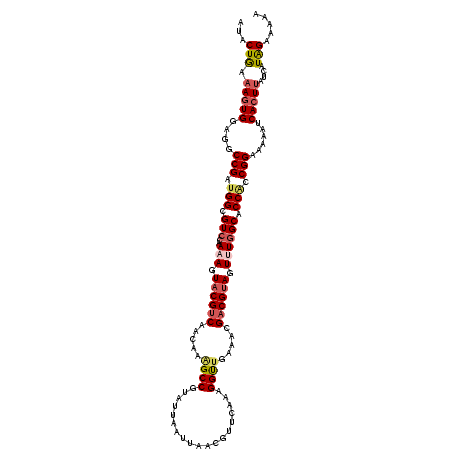

| Location | 15,886,424 – 15,886,544 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.83 |
| Mean single sequence MFE | -41.72 |
| Consensus MFE | -30.94 |
| Energy contribution | -31.58 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15886424 120 + 22407834 UUGAUGGAGUCUGUCACCGGCAUAUUUCGAAAUGGCGGUGAUUUUCAGAGUAGUCAGGCCCACUUUCUUUGGGCGGAUCAUGAAUGAAAUGCCACGUCCUGUAUUCGCGGGAAUCCAUAA ...(((((....(((((((.(((........))).)))))))(((((..((.(((..(((((.......))))).))).))...))))).......((((((....)))))).))))).. ( -40.00) >DroSec_CAF1 59866 120 + 1 UUGAUGGAGUCUGUCACCGGCAUAUGACGAGAUGGCGGUGAUUUUCAGAGUGGUCAGACCCAUUUUCUUUGGGCGGAUCAUGAAUGAAAUGCCACGUCCCGUAUUUGCGGGAAUCCAUAA ...(((((....(((((((.(((........))).)))))))(((((..((((((.(.((((.......))))).))))))...))))).......((((((....)))))).))))).. ( -45.30) >DroSim_CAF1 60062 120 + 1 UUGAUGGAGUCUGUCACCGGCAUAUGACGAAAUGGCGGUGAUUUUCAGAGUGGUCAGACCCACUUUCUUUGGGCGGAUCAUGAAUGAAAUGCCACGUCCCGUAUUUGCGGGAAUCCAUAA ...(((((....(((((((.(((........))).)))))))(((((..((((((.(.((((.......))))).))))))...))))).......((((((....)))))).))))).. ( -44.30) >DroEre_CAF1 49426 120 + 1 UUGAUGGAGUCUGUCGCCGGCAUUUGGCGAGAUGGCGGAGAUCUUUAGGGUGGUCAGACCUACUUUCCUCGGGCGGAUCAUGAAGGAGAUGCUUCGUCCGGUACUGGCGGGUAUCCAUAA ...((((((((((((((((.(((((....))))).)((....))....))))).))))).((((..(((((((((((.(((.......))).)))))))))....))..))))))))).. ( -40.80) >DroYak_CAF1 37784 120 + 1 UUGAUUUAGUCUGUCACCGGCAAAUGGCGAGAUGGCGGUGAUCUUCAGAGCGGUCAGACCAACUUUCUUCGGGCGGAUCAUGAACGAUAUGCUACGUCCGGUAUUUGCCGGUAUCCAUAU ..(((.......)))(((((((((((.((.((((..(((((((((((((..((.....))..((.((....)).)).)).)))).))).)))).)))))).)))))))))))........ ( -38.20) >consensus UUGAUGGAGUCUGUCACCGGCAUAUGACGAGAUGGCGGUGAUUUUCAGAGUGGUCAGACCCACUUUCUUUGGGCGGAUCAUGAAUGAAAUGCCACGUCCCGUAUUUGCGGGAAUCCAUAA ...((((((...(((((((.(((........))).)))))))...)(((((((......)))))))...((((((..((......))..)))).))((((((....)))))).))))).. (-30.94 = -31.58 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:33 2006