| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,881,644 – 15,881,746 |
| Length | 102 |
| Max. P | 0.761055 |

| Location | 15,881,644 – 15,881,746 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.77 |
| Mean single sequence MFE | -22.30 |
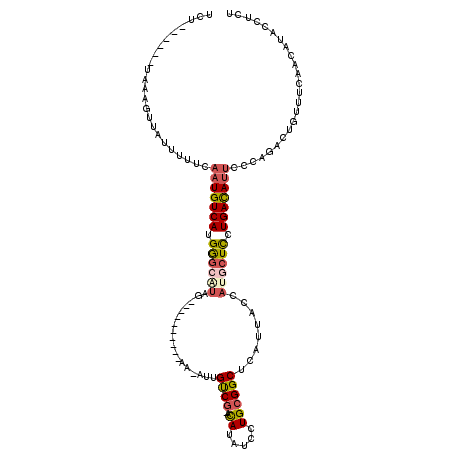
| Consensus MFE | -9.18 |
| Energy contribution | -9.82 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
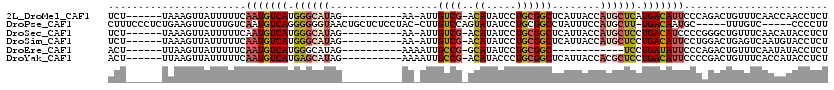
>2L_DroMel_CAF1 15881644 102 + 22407834 UCU------UAAAGUUAUUUUUCAAUGUCAUGGGCAUAG----------AA-AUUGUCG-ACAUAUCCUGCGGCUCAUUACCAUGCUCAUGACAUUCCCAGACUGUUUCAACCAACCUCU ...------...((((.......((((((((((((((.(----------.(-((.((((-.((.....))))))..))).).))))))))))))))....))))................ ( -24.90) >DroPse_CAF1 47935 108 + 1 CUUUCCCUCUGAAGUUCUUUGUCAAUGUCAGGGGGGUAACUGCUCUCCUAC-CUUGUCCAGUAUAUCCUGCGGCCUAUUUCCAUGCUU-UGACAAUGC-----UUUGUC-----CCCCUU ..........(((((...(((((((.((.((((((((....))))))))..-...((((((......))).)))..........)).)-)))))).))-----)))...-----...... ( -26.30) >DroSec_CAF1 55117 102 + 1 UCU------UAAAGUUAUUUUUCAAUGUCAUGGGCAUAG----------AA-AUUGUCG-ACAUAUCCUGCGGCUCAUUACCAUGCUCCUGACAUCCCCGGGCUGUUUCAACAUACCUCU ...------...............((((((.((((((.(----------.(-((.((((-.((.....))))))..))).).)))))).))))))....((..(((....)))..))... ( -22.00) >DroSim_CAF1 55208 102 + 1 UCU------UAAAGUUAUUUUUCAAUGUCAUGGGCAUAG----------AA-AUUGUCG-ACAUAUCCUGCGGCUCAUUACCAUGCUCCUGACAUUCCUGGACUGAGUCAAUGUACCUCU ...------...((((.......(((((((.((((((.(----------.(-((.((((-.((.....))))))..))).).)))))).)))))))....))))(((.........))). ( -22.00) >DroEre_CAF1 44604 91 + 1 ACU------UUAAGUUAUUUUUCAAUGUCAUGGGCAUAG----------AAAAUUGCCG-GCAUAUCCUGCGGC------------UCCUGAUAUUCCCAGACUGUUUCAAUAUACCUCU ...------.................(((.((((.(((.----------......((((-.((.....))))))------------......))).)))))))................. ( -17.42) >DroYak_CAF1 32179 103 + 1 ACU------UUAAGUUAUUUUUCAAUGUCAUGAGCAUAG----------AAAAUUGCCG-ACAUACCCUGCGGCUCAUUACCACGCUCCUGACAUUCCCCGACUGUUUCACCAUACCUCU ...------...((((.......(((((((.((((....----------......((((-.((.....))))))..........)))).)))))))....))))................ ( -21.15) >consensus UCU______UAAAGUUAUUUUUCAAUGUCAUGGGCAUAG__________AA_AUUGUCG_ACAUAUCCUGCGGCUCAUUACCAUGCUCCUGACAUUCCCAGACUGUUUCAACAUACCUCU .......................(((((((.((((((..................((((..((.....))))))........)))))).)))))))........................ ( -9.18 = -9.82 + 0.64)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:28 2006