
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,877,990 – 15,878,132 |
| Length | 142 |
| Max. P | 0.999874 |

| Location | 15,877,990 – 15,878,092 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -35.18 |
| Consensus MFE | -31.18 |
| Energy contribution | -31.46 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.89 |
| SVM decision value | 4.33 |
| SVM RNA-class probability | 0.999874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15877990 102 - 22407834 AAUUUGCAAUGGAUUUUGGGGGCGAAGAGCUGGAAAACUUAAUUAGCCAGCCGCAGUCAGAUAUCCAAUUGCGAUGGUGAUUGUUCCUGAAACCGAGGCAAA ..(((((..(((.(((..(((((((...(((((..((.....))..)))))((((((..(....)..)))))).......)))))))..))))))..))))) ( -33.90) >DroSec_CAF1 51491 102 - 1 AAUUUGCACUGGAUUUUGGGGGCGAAGAGCUGGAAAACUUAAUUACCCAGCCGCAGUCCGAUAUCCAAUUGGGAUGGUGAUUGUUCCUGAAACCGAGGCAAA ..(((((..(((.(((..(((((((...(((((.............)))))(((..((((((.....))))))...))).)))))))..))))))..))))) ( -35.72) >DroSim_CAF1 51560 102 - 1 AAUUUGCACUGGAUUUUGGGGGCGAAGAGCUGGAAAACUUAAUUAGCCAGCCGCAGUCCGAUAUCCAAUUGGGAUGGUGAUUGUUCCUGAAACCGAGGCAAA ..(((((..(((.(((..(((((((...(((((..((.....))..)))))(((..((((((.....))))))...))).)))))))..))))))..))))) ( -35.70) >DroEre_CAF1 41555 102 - 1 AAUUUGCAAUGGAUUUCGGGGGCGUGGAGCUGGAAAACUUAAUUAGCCAGCCGCAGUCCAAAAUCCAAAUGGGAUGGUGAUUGUUCCUGAAACCGAGGCAAA ..(((((..(((.(((((((((((((..(((((..((.....))..))))))))(((((...((((.....)))).).)))))))))))))))))..))))) ( -36.30) >DroYak_CAF1 27746 102 - 1 AAUUUUCAAUGGAUUUUGGGGGCGAAGAGCUGGAAAACUUAAUUAGCCAGCCGCAAUCCCAAAUCCAAUUGGGAUGGUGAUUGUUCCUGAAACCGAGGCAAA .........(((.(((..(((((((...(((((..((.....))..)))))(((.(((((((......))))))).))).)))))))..))))))....... ( -34.30) >consensus AAUUUGCAAUGGAUUUUGGGGGCGAAGAGCUGGAAAACUUAAUUAGCCAGCCGCAGUCCGAUAUCCAAUUGGGAUGGUGAUUGUUCCUGAAACCGAGGCAAA ..(((((..(((.((((((((((((...(((((.............)))))(((.((((............)))).))).)))))))))))))))..))))) (-31.18 = -31.46 + 0.28)

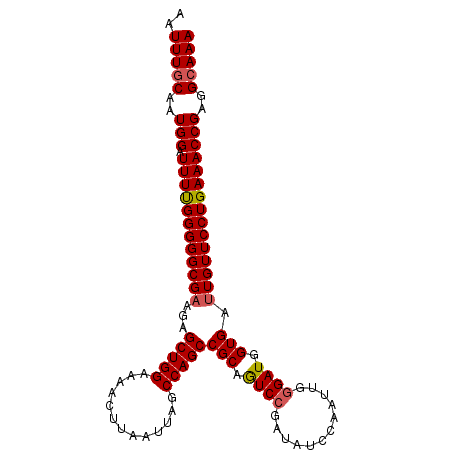

| Location | 15,878,025 – 15,878,132 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -26.69 |
| Consensus MFE | -17.65 |
| Energy contribution | -17.91 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15878025 107 + 22407834 UGGAUAUCUGACUGCGGCUGGCUAAUUAAGUUUUCCAGCUCUUCGCCCCCAAAAUCCAUUGCAAAUUGGCCCCUUAAAUUUGCCUAGGCCGCAAAAUUUUCCACCAU ((((........((((((((((......(((......)))....))).............(((((((.........)))))))...)))))))......)))).... ( -24.04) >DroSec_CAF1 51526 107 + 1 UGGAUAUCGGACUGCGGCUGGGUAAUUAAGUUUUCCAGCUCUUCGCCCCCAAAAUCCAGUGCAAAUUGGCCCCUAAAAUUUUCCUACGCCGCAAAAUUUUCCACCAU ((((........((((((.(((..(...(((......)))..)..))).......(((((....)))))..................))))))......)))).... ( -24.14) >DroSim_CAF1 51595 107 + 1 UGGAUAUCGGACUGCGGCUGGCUAAUUAAGUUUUCCAGCUCUUCGCCCCCAAAAUCCAGUGCAAAUUGGCCCCUAAAAUUUGCCUAGGCCGCAAAAUUUUCCACCAU ((((........((((((((((......(((......)))....)))..........((.(((((((.........))))))))).)))))))......)))).... ( -25.34) >DroEre_CAF1 41590 107 + 1 UGGAUUUUGGACUGCGGCUGGCUAAUUAAGUUUUCCAGCUCCACGCCCCCGAAAUCCAUUGCAAAUUGGCCCCUAAAAUUUGCCUAGGCCACAAAAUUUUCCACCAU ((((((((((...((((((((..(((...)))..)))))....)))..))))))))))........(((((...............)))))................ ( -28.96) >DroYak_CAF1 27781 107 + 1 UGGAUUUGGGAUUGCGGCUGGCUAAUUAAGUUUUCCAGCUCUUCGCCCCCAAAAUCCAUUGAAAAUUGGCCCCUAAAACUUGCCUAGGCCGCAAAAUUUUCCACCAU ((((((((((...((((((((..(((...)))..)))))....))).))).)))))))..(((((((((((...............))))....)))))))...... ( -28.96) >DroPer_CAF1 70675 83 + 1 UGGAUGUUGCGCUGCGGCAGGCUAAUUAAGCUGU--------GUGCCA-------------CAGAAUGGUC--U-AAAAUUGUUUAUGCCGCAGCAUUUUCCAAUAU ((((((((((.(((.((((.((..........))--------.)))).-------------)))...(((.--(-(((....)))).)))))))))...)))).... ( -28.70) >consensus UGGAUAUCGGACUGCGGCUGGCUAAUUAAGUUUUCCAGCUCUUCGCCCCCAAAAUCCAUUGCAAAUUGGCCCCUAAAAUUUGCCUAGGCCGCAAAAUUUUCCACCAU ((((........((((((((((.((((.(((......)))....(((....................)))......)))).)))..)))))))......)))).... (-17.65 = -17.91 + 0.25)

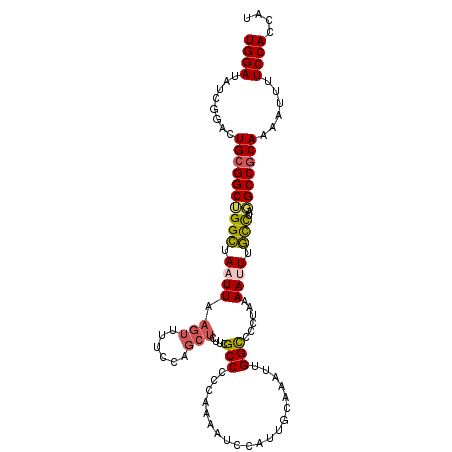
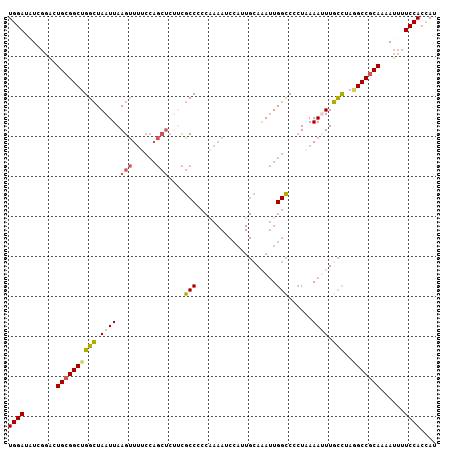
| Location | 15,878,025 – 15,878,132 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -29.81 |
| Consensus MFE | -18.65 |
| Energy contribution | -19.88 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15878025 107 - 22407834 AUGGUGGAAAAUUUUGCGGCCUAGGCAAAUUUAAGGGGCCAAUUUGCAAUGGAUUUUGGGGGCGAAGAGCUGGAAAACUUAAUUAGCCAGCCGCAGUCAGAUAUCCA ....((((.....(((((((...(((....(((((...(((.(((((..(.(....).)..)))))....)))....)))))...))).))))))).......)))) ( -28.40) >DroSec_CAF1 51526 107 - 1 AUGGUGGAAAAUUUUGCGGCGUAGGAAAAUUUUAGGGGCCAAUUUGCACUGGAUUUUGGGGGCGAAGAGCUGGAAAACUUAAUUACCCAGCCGCAGUCCGAUAUCCA .((((..((((((((.(......).))))))))....)))).........((((.(((((.(((....(((((.............))))))))..))))).)))). ( -30.22) >DroSim_CAF1 51595 107 - 1 AUGGUGGAAAAUUUUGCGGCCUAGGCAAAUUUUAGGGGCCAAUUUGCACUGGAUUUUGGGGGCGAAGAGCUGGAAAACUUAAUUAGCCAGCCGCAGUCCGAUAUCCA ..((((..(((((....(((((.............)))))))))).))))((((.(((((.(((....(((((..((.....))..))))))))..))))).)))). ( -31.52) >DroEre_CAF1 41590 107 - 1 AUGGUGGAAAAUUUUGUGGCCUAGGCAAAUUUUAGGGGCCAAUUUGCAAUGGAUUUCGGGGGCGUGGAGCUGGAAAACUUAAUUAGCCAGCCGCAGUCCAAAAUCCA ....((((........((((((.............))))))........((((((.(((.(((...(((.(....).))).....)))..))).))))))...)))) ( -30.72) >DroYak_CAF1 27781 107 - 1 AUGGUGGAAAAUUUUGCGGCCUAGGCAAGUUUUAGGGGCCAAUUUUCAAUGGAUUUUGGGGGCGAAGAGCUGGAAAACUUAAUUAGCCAGCCGCAAUCCCAAAUCCA ......(((((((....(((((.............))))))))))))..(((((((.(((.(((....(((((..((.....))..))))))))...)))))))))) ( -32.22) >DroPer_CAF1 70675 83 - 1 AUAUUGGAAAAUGCUGCGGCAUAAACAAUUUU-A--GACCAUUCUG-------------UGGCAC--------ACAGCUUAAUUAGCCUGCCGCAGCGCAACAUCCA ....((((...((((((((((...........-.--(.((((...)-------------))))..--------...(((.....))).)))))))))).....)))) ( -25.80) >consensus AUGGUGGAAAAUUUUGCGGCCUAGGCAAAUUUUAGGGGCCAAUUUGCAAUGGAUUUUGGGGGCGAAGAGCUGGAAAACUUAAUUAGCCAGCCGCAGUCCGAUAUCCA ....((((.....(((((((...(((.((((.......(((.(((((..............)))))....))).......)))).))).))))))).......)))) (-18.65 = -19.88 + 1.23)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:24 2006