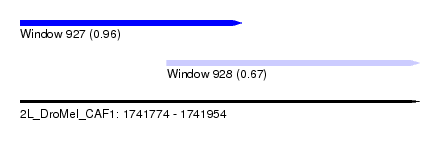
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,741,774 – 1,741,954 |
| Length | 180 |
| Max. P | 0.955205 |

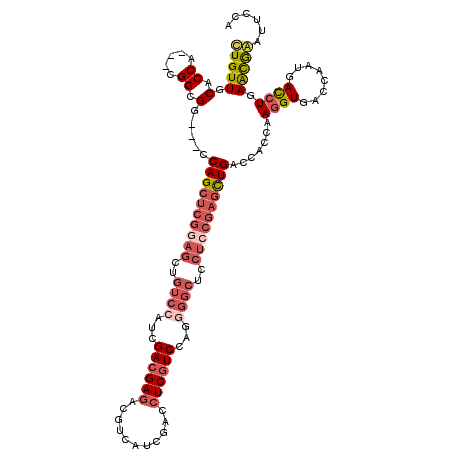
| Location | 1,741,774 – 1,741,874 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.18 |
| Mean single sequence MFE | -49.80 |
| Consensus MFE | -18.69 |
| Energy contribution | -19.75 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1741774 100 + 22407834 CAG------------UUCCGGCGUCUGCAGCCUCAAGGGUCACCAUGGUG--ACCCCGACAGCGGUUGCCUGGGCAUCAACUGUUGCACCA---CGGCGG---CCAGCUCGGAGCUGUCC (((------------(((((((.......((.....(((((((....)))--)))).....))(((((((((((((........))).)))---.)))))---)).)).))))))))... ( -52.40) >DroSim_CAF1 5390 100 + 1 CAG------------UUCCGGCGUCUGCAGCCUCAAGGGUCACCAUGGUG--ACCCCGACGGCGGUUGCCUGGGCAUCAACUGUUGCACCA---CGGCGG---CCAGCUCGGAGCUGUCC (((------------(((((((.......(((((..(((((((....)))--)))).)).)))(((((((((((((........))).)))---.)))))---)).)).))))))))... ( -56.60) >DroWil_CAF1 14708 118 + 1 CAGUGGAAGUGGAAGUUGCGGCAAUGUCAACACGAAAGGACAACUUCAUGAGAGCACGAA--AGGAUGCCUGGGCUUUGAUUGUUGCACCGCCAAGGCGGGAUCCAGUUCCGAAAUAUCC ...(((((.((((...((((((((((((....(....)))).........(((((.....--(((...)))..))))).)))))))))((((....))))..)))).)))))........ ( -38.90) >DroYak_CAF1 5407 100 + 1 CAG------------UUCCGGCGUCGGUAGUCUCAAGGAUCACCAUGGUG--ACCACGACGGCGGCUGCCUGGGCAUCAACUGUUGCACCA---CGGCGG---CCAGCUCGGAGCUGUCC (((------------(((((((((((...((((((.((....)).))).)--))..))))...(((((((((((((........))).)))---.)))))---)).)).))))))))... ( -51.30) >consensus CAG____________UUCCGGCGUCGGCAGCCUCAAGGGUCACCAUGGUG__ACCACGACGGCGGUUGCCUGGGCAUCAACUGUUGCACCA___CGGCGG___CCAGCUCGGAGCUGUCC ...............(((((((..............((..(((....)))...))........(((((((((((((........))).)))....)))))...)).)).)))))...... (-18.69 = -19.75 + 1.06)

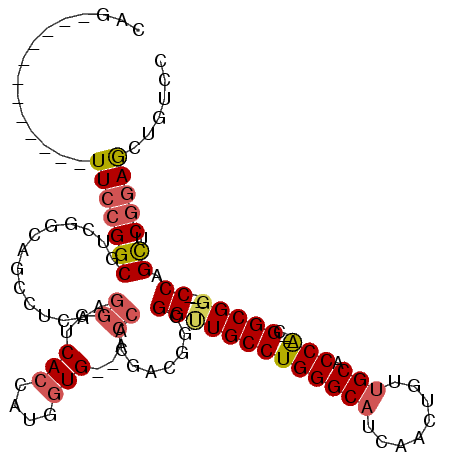
| Location | 1,741,840 – 1,741,954 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.07 |
| Mean single sequence MFE | -43.47 |
| Consensus MFE | -24.10 |
| Energy contribution | -27.10 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1741840 114 + 22407834 CUGUUGCACCA---CGGCGG---CCAGCUCGGAGCUGUCCAUCGACGAGACGUCAUCGACCUCGUCCAGAGGCUCCUGCGAGUUGACCAGCAAGGUGACCAACGACCUGAACGGAUUCCA .(((((((((.---..((((---.(((((((.((..(((..(.((((((.((....))..)))))).)..)))..)).))))))).)).))..))))..))))).((.....))...... ( -40.90) >DroSim_CAF1 5456 114 + 1 CUGUUGCACCA---CGGCGG---CCAGCUCGGAGCUGUCCAUCGACGAGACGUCAUCGACCUCGUCCAGGGGCUCCUCCGAGCUGACCAGCAAGGUGACCAAUGACCUGAACGGAUUCCA (((((.((((.---..((((---.((((((((((..((((...((((((.((....))..))))))...))))..)))))))))).)).))..))))..((......)))))))...... ( -50.90) >DroWil_CAF1 14786 111 + 1 UUGUUGCACCGCCAAGGCGGGAUCCAGUUCCGAAAUAUCCAUCGAUGAGACUGCAUCGACAUCUUCGCG---------CACUCUGGCAGCGAAAGUGAACAAUAAUCUGAGUAAUUUCGA ........((((....))))..........((((((.....((((((......))))))((..((((((---------(......)).)))))..))................)))))). ( -30.40) >DroYak_CAF1 5473 114 + 1 CUGUUGCACCA---CGGCGG---CCAGCUCGGAGCUGUCCAUCGACGAGACGUCGUCGACCUCGUCCCGGGGCUCCUCCGAGCUGACCACCAAGGUGACCAAUGACCUGAGCGAAUUCCA ...((((....---.((.((---.((((((((((..((((...((((((.((....))..))))))...))))..)))))))))).)).)).((((........))))..))))...... ( -51.70) >consensus CUGUUGCACCA___CGGCGG___CCAGCUCGGAGCUGUCCAUCGACGAGACGUCAUCGACCUCGUCCAGGGGCUCCUCCGAGCUGACCACCAAGGUGACCAAUGACCUGAACGAAUUCCA (((((.(.((.....)).).....((((((((((..((((...((((((...........))))))...))))..)))))))))).......((((........)))).)))))...... (-24.10 = -27.10 + 3.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:44 2006