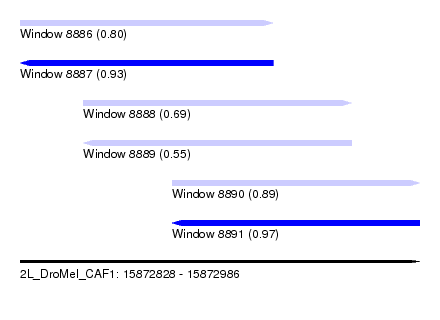
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,872,828 – 15,872,986 |
| Length | 158 |
| Max. P | 0.970870 |

| Location | 15,872,828 – 15,872,928 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.30 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -14.29 |
| Energy contribution | -14.43 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
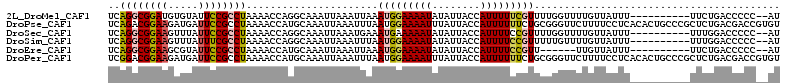
>2L_DroMel_CAF1 15872828 100 + 22407834 AU--GGGGGUCAGAA----------AAAUAACAAAACCAAAACGAAAAAUGGUAAUAUAUUUUCCAUUUAAUUUAAUUUGCCUGGUUUUAGGCGGAAUACACAUCCGCCUGA ..--...........----------.......(((((((...((((((((((.((.....)).)))))).......))))..)))))))(((((((.......))))))).. ( -21.11) >DroPse_CAF1 38450 112 + 1 ACACGGUCGUCAGAGCGGGCAGUGUGAGGAAAAGAACCCGCAGAAAAAAUGGUAAUAAAUUUUCCAUUAAAUUUAAUUUGCAUGGUUUUAGGCGGAAUCAUCUUCCGUCUGA (((((.((((....)))).).)))).......((((((.(((((((((((((.((.....)).)))))...)))..)))))..))))))((((((((.....)))))))).. ( -28.00) >DroSec_CAF1 46303 100 + 1 AU--GGGGGUCCAAA----------AAAUAACAAAACCAAAACGGAAAAUGGUAAUAUAUUUUUCAUUUCAUUUAAUUUGCCUGGUUUUAGGCGGAAUAAACUUCCGCCUGA .(--((....)))..----------.......(((((((....((((((((......))))))))....((.......))..)))))))((((((((.....)))))))).. ( -24.00) >DroSim_CAF1 46378 100 + 1 AU--GGGGGUCCAAA----------AAAUAACAAAACAAAAACGGAAAAUGGUAAUAUAUUUUCCAUUAAAUUUAAUUUGCCUGGUUUUAGGCGAAAUAAACUUCCGCCUGA ..--((((((.....----------..................((((((((......)))))))).......(((.((((((((....)))))))).)))))))))...... ( -23.40) >DroEre_CAF1 36367 94 + 1 AU--GGGGGUCAGAA----------AAAUAACAA------AACGGAAAAUGGUAAUAUAUUUUCCAUUUAAUUUAAUUUGCAUGGUUUUAGGCGGAAUACGCUUCCGCCUGA ..--.(((..((..(----------((.(((..(------((.((((((((......)))))))).)))...))).)))...))..)))((((((((.....)))))))).. ( -22.50) >DroPer_CAF1 64175 112 + 1 ACACGGUCGUCAGAGCGGGCAGUGUGAGGAAAAGAACCCGCAGAAAAAAUGGUAAUAAAUUUUCCAUUAAAUUUAAUUUGCAUGGUUUUAGGCGGAAUCAUCUUCCGUCCGA ...(((.(((((((((.(((((((((.((.......)))))).....(((((.((.....)).))))).........)))).).))))).)))((((.....))))).))). ( -26.80) >consensus AU__GGGGGUCAGAA__________AAAUAACAAAACCAAAACGAAAAAUGGUAAUAUAUUUUCCAUUAAAUUUAAUUUGCAUGGUUUUAGGCGGAAUAAACUUCCGCCUGA ...............................................(((((.((.....)).)))))...................((((((((((.....)))))))))) (-14.29 = -14.43 + 0.14)


| Location | 15,872,828 – 15,872,928 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.30 |
| Mean single sequence MFE | -23.53 |
| Consensus MFE | -13.78 |
| Energy contribution | -14.12 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15872828 100 - 22407834 UCAGGCGGAUGUGUAUUCCGCCUAAAACCAGGCAAAUUAAAUUAAAUGGAAAAUAUAUUACCAUUUUUCGUUUUGGUUUUGUUAUUU----------UUCUGACCCCC--AU ..(((((((.......))))))).....((((.((((.((((((((((((((((........)))))))).))))))))....))))----------.))))......--.. ( -24.40) >DroPse_CAF1 38450 112 - 1 UCAGACGGAAGAUGAUUCCGCCUAAAACCAUGCAAAUUAAAUUUAAUGGAAAAUUUAUUACCAUUUUUUCUGCGGGUUCUUUUCCUCACACUGCCCGCUCUGACGACCGUGU ((((((((((.....))))).......((((..((((...)))).))))......................((((((...............)))))))))))......... ( -22.16) >DroSec_CAF1 46303 100 - 1 UCAGGCGGAAGUUUAUUCCGCCUAAAACCAGGCAAAUUAAAUGAAAUGAAAAAUAUAUUACCAUUUUCCGUUUUGGUUUUGUUAUUU----------UUUGGACCCCC--AU ..((((((((.....))))))))(((((((((......(((((..................))))).....))))))))).......----------..(((....))--). ( -22.77) >DroSim_CAF1 46378 100 - 1 UCAGGCGGAAGUUUAUUUCGCCUAAAACCAGGCAAAUUAAAUUUAAUGGAAAAUAUAUUACCAUUUUCCGUUUUUGUUUUGUUAUUU----------UUUGGACCCCC--AU ..((((((((.....))))))))....(((((.((((.((((..((((((((((........))))))))))...))))....))))----------)))))......--.. ( -24.90) >DroEre_CAF1 36367 94 - 1 UCAGGCGGAAGCGUAUUCCGCCUAAAACCAUGCAAAUUAAAUUAAAUGGAAAAUAUAUUACCAUUUUCCGUU------UUGUUAUUU----------UUCUGACCCCC--AU ..((((((((.....)))))))).....((.(.(((.(((...(((((((((((........))))))))))------)..))).))----------).)))......--.. ( -24.20) >DroPer_CAF1 64175 112 - 1 UCGGACGGAAGAUGAUUCCGCCUAAAACCAUGCAAAUUAAAUUUAAUGGAAAAUUUAUUACCAUUUUUUCUGCGGGUUCUUUUCCUCACACUGCCCGCUCUGACGACCGUGU ..((.(((((.....)))))))......((((...............((((((............))))))((((((...............)))))).........)))). ( -22.76) >consensus UCAGGCGGAAGUUUAUUCCGCCUAAAACCAGGCAAAUUAAAUUAAAUGGAAAAUAUAUUACCAUUUUCCGUUUUGGUUUUGUUAUUU__________UUCUGACCCCC__AU ..((((((((.....))))))))......................(((((((((........)))))))))......................................... (-13.78 = -14.12 + 0.33)


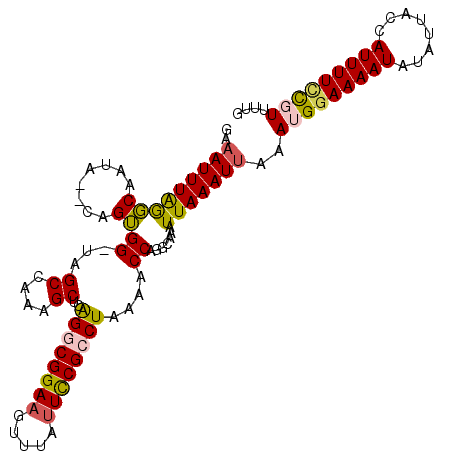
| Location | 15,872,853 – 15,872,959 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -17.54 |
| Energy contribution | -18.02 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15872853 106 + 22407834 CAAAACGAAAAAUGGUAAUAUAUUUUCCAUUUAAUUUAAUUUGCCUGGUUUUAGGCGGAAUACACAUCCGCCUGAGCUUUGGCUA-CCACCG--CACUGCCUAAAUUUC .....((..((((((.((.....)).))))))..........(((.(((((.(((((((.......))))))))))))..)))..-....))--............... ( -25.10) >DroPse_CAF1 38487 109 + 1 CCGCAGAAAAAAUGGUAAUAAAUUUUCCAUUAAAUUUAAUUUGCAUGGUUUUAGGCGGAAUCAUCUUCCGUCUGAGCGCUGGCCAGCCCCUGUUUGUGGGUUAAAUUUC (((((((.....((((..(((((((......)))))))........((((((((((((((.....))))))))))).))).)))).......))))))).......... ( -29.90) >DroSec_CAF1 46328 106 + 1 CAAAACGGAAAAUGGUAAUAUAUUUUUCAUUUCAUUUAAUUUGCCUGGUUUUAGGCGGAAUAAACUUCCGCCUGAGCUUUGGCGG-CCACUG--CAUUGCCUAAAUUUC ......((((((((......))))))))..............(((.(((((.((((((((.....)))))))))))))..)))((-(.....--....)))........ ( -30.30) >DroSim_CAF1 46403 106 + 1 AAAAACGGAAAAUGGUAAUAUAUUUUCCAUUAAAUUUAAUUUGCCUGGUUUUAGGCGAAAUAAACUUCCGCCUGAGCUUUGGCGC-CCACUG--CAUUGCCUAAAUUUC ......((((((((......))))))))...(((((((....(((.(((((.(((((...........))))))))))..)))((-.....)--)......))))))). ( -26.40) >DroEre_CAF1 36389 103 + 1 ---AACGGAAAAUGGUAAUAUAUUUUCCAUUUAAUUUAAUUUGCAUGGUUUUAGGCGGAAUACGCUUCCGCCUGAGCUUUGGCUC-CCACGG--UAUUGCCCAAAUUUC ---...((((((((......)))))))).........((((((..(((....((((((((.....))))))))((((....))))-))).((--.....)))))))).. ( -30.60) >DroPer_CAF1 64212 109 + 1 CCGCAGAAAAAAUGGUAAUAAAUUUUCCAUUAAAUUUAAUUUGCAUGGUUUUAGGCGGAAUCAUCUUCCGUCCGAGCGCUGGCCAGCCCCUGUUUGUGGGUUAAAUUUC (((((((.....((((..(((((((......)))))))........(((((..(((((((.....)))))))..)).))).)))).......))))))).......... ( -26.40) >consensus CAAAACGAAAAAUGGUAAUAUAUUUUCCAUUAAAUUUAAUUUGCAUGGUUUUAGGCGGAAUAAACUUCCGCCUGAGCUUUGGCCA_CCACUG__CAUUGCCUAAAUUUC ..........(((((.((.....)).)))))...........((..(((((.((((((((.....)))))))))))))...)).......................... (-17.54 = -18.02 + 0.47)

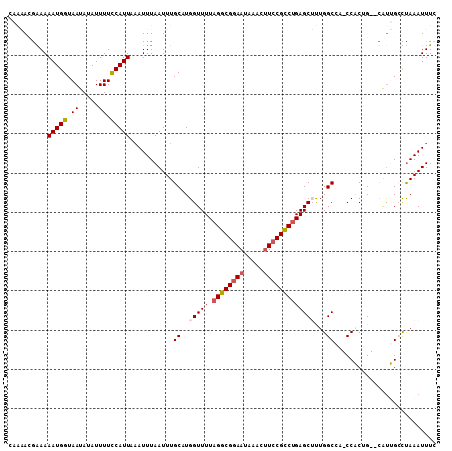
| Location | 15,872,853 – 15,872,959 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.62 |
| Mean single sequence MFE | -26.68 |
| Consensus MFE | -16.58 |
| Energy contribution | -16.28 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15872853 106 - 22407834 GAAAUUUAGGCAGUG--CGGUGG-UAGCCAAAGCUCAGGCGGAUGUGUAUUCCGCCUAAAACCAGGCAAAUUAAAUUAAAUGGAAAAUAUAUUACCAUUUUUCGUUUUG ..(((((((....((--(..(((-((((....))).(((((((.......)))))))...)))).)))..)))))))(((((((((((........))))))))))).. ( -29.00) >DroPse_CAF1 38487 109 - 1 GAAAUUUAACCCACAAACAGGGGCUGGCCAGCGCUCAGACGGAAGAUGAUUCCGCCUAAAACCAUGCAAAUUAAAUUUAAUGGAAAAUUUAUUACCAUUUUUUCUGCGG .(((((((((((.......)))(((((...(((.(((..(....).)))...)))......))).))...))))))))...((((((............)))))).... ( -18.80) >DroSec_CAF1 46328 106 - 1 GAAAUUUAGGCAAUG--CAGUGG-CCGCCAAAGCUCAGGCGGAAGUUUAUUCCGCCUAAAACCAGGCAAAUUAAAUGAAAUGAAAAAUAUAUUACCAUUUUCCGUUUUG ...((((((((....--.....)-))(((.......((((((((.....)))))))).......)))....)))))((((((.(((((........))))).)))))). ( -26.34) >DroSim_CAF1 46403 106 - 1 GAAAUUUAGGCAAUG--CAGUGG-GCGCCAAAGCUCAGGCGGAAGUUUAUUUCGCCUAAAACCAGGCAAAUUAAAUUUAAUGGAAAAUAUAUUACCAUUUUCCGUUUUU .((((((((....((--(...((-((......))))((((((((.....))))))))........)))..))))))))((((((((((........))))))))))... ( -30.60) >DroEre_CAF1 36389 103 - 1 GAAAUUUGGGCAAUA--CCGUGG-GAGCCAAAGCUCAGGCGGAAGCGUAUUCCGCCUAAAACCAUGCAAAUUAAAUUAAAUGGAAAAUAUAUUACCAUUUUCCGUU--- ..((((((((.....--))((((-((((....))))((((((((.....))))))))....)))).))))))......((((((((((........))))))))))--- ( -34.80) >DroPer_CAF1 64212 109 - 1 GAAAUUUAACCCACAAACAGGGGCUGGCCAGCGCUCGGACGGAAGAUGAUUCCGCCUAAAACCAUGCAAAUUAAAUUUAAUGGAAAAUUUAUUACCAUUUUUUCUGCGG .(((((((((((.......)))(((((.........((.(((((.....))))))).....))).))...))))))))...((((((............)))))).... ( -20.54) >consensus GAAAUUUAGGCAAUA__CAGUGG_UAGCCAAAGCUCAGGCGGAAGUUUAUUCCGCCUAAAACCAGGCAAAUUAAAUUAAAUGGAAAAUAUAUUACCAUUUUCCGUUUUG ..(((((((((........))((...((....))..((((((((.....))))))))....)).......)))))))..(((((((((........))))))))).... (-16.58 = -16.28 + -0.30)

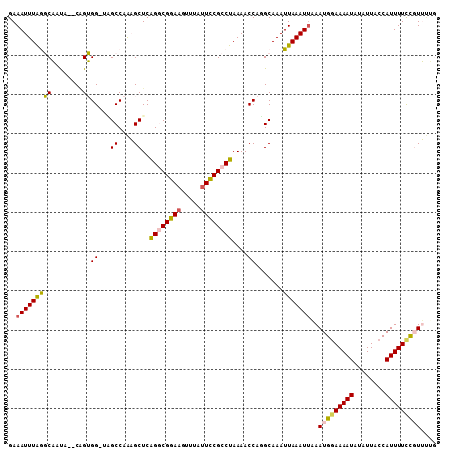
| Location | 15,872,888 – 15,872,986 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 79.61 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -17.07 |
| Energy contribution | -16.90 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.893054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15872888 98 + 22407834 UUAAUUUGCCUGGUUUUAGGCGGAAUACACAUCCGCCUGAGCUUUGGCUA-CCACCG--CACUGCCUAAAUUUCAUUAGGCGUAAAGCAAACUUU-UUUGUU .......(((.(((((.(((((((.......))))))))))))..)))..-.....(--(..(((((((......)))))))....)).......-...... ( -29.90) >DroPse_CAF1 38522 102 + 1 UUAAUUUGCAUGGUUUUAGGCGGAAUCAUCUUCCGUCUGAGCGCUGGCCAGCCCCUGUUUGUGGGUUAAAUUUCAUUAUGCGUGAAAGAUGCUUUCCUUAUU .......((((((((((((((((((.....))))))))))).....)))((((((.....).)))))...((((((.....)))))).)))).......... ( -27.50) >DroSec_CAF1 46363 98 + 1 UUAAUUUGCCUGGUUUUAGGCGGAAUAAACUUCCGCCUGAGCUUUGGCGG-CCACUG--CAUUGCCUAAAUUUCAUUUAGCGCAAAGCAAACUUU-UUUGUU .....(((((.(((((.((((((((.....)))))))))))))..)))))-....((--(.((((((((((...)))))).)))).)))......-...... ( -33.80) >DroSim_CAF1 46438 98 + 1 UUAAUUUGCCUGGUUUUAGGCGAAAUAAACUUCCGCCUGAGCUUUGGCGC-CCACUG--CAUUGCCUAAAUUUCAUUCAAAGCAAAGCAAACUUU-UUUGUU ....((((..(((.(((((((((...........(((........)))((-.....)--).)))))))))..)))..))))....((((((....-)))))) ( -21.50) >DroEre_CAF1 36421 97 + 1 UUAAUUUGCAUGGUUUUAGGCGGAAUACGCUUCCGCCUGAGCUUUGGCUC-CCACGG--UAUUGCCCAAAUUUCAUUAGGCGUAAAACGAACUUU--UUGCU .......(((.(((((.((((((((.....))))))))((((....))))-.....(--(.((((((...........)).)))).)))))))..--.))). ( -27.50) >DroPer_CAF1 64247 102 + 1 UUAAUUUGCAUGGUUUUAGGCGGAAUCAUCUUCCGUCCGAGCGCUGGCCAGCCCCUGUUUGUGGGUUAAAUUUCAUUAUGCGUGAAAGAUGCUUUCCUUAUU .......(((((((((..(((((((.....)))))))..)).....)))((((((.....).)))))...((((((.....)))))).)))).......... ( -24.00) >consensus UUAAUUUGCAUGGUUUUAGGCGGAAUAAACUUCCGCCUGAGCUUUGGCCA_CCACUG__CAUUGCCUAAAUUUCAUUAAGCGUAAAACAAACUUU_UUUGUU ....(((((......((((((((((.....))))))))))((....)).................................)))))................ (-17.07 = -16.90 + -0.17)

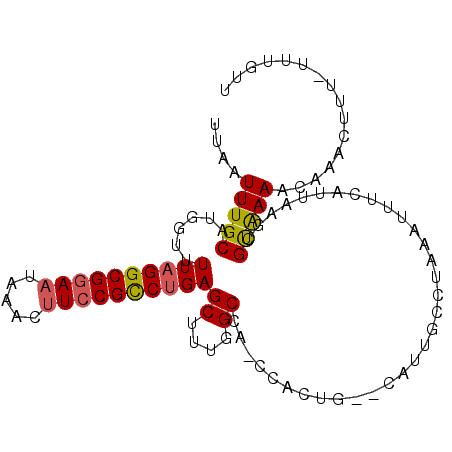
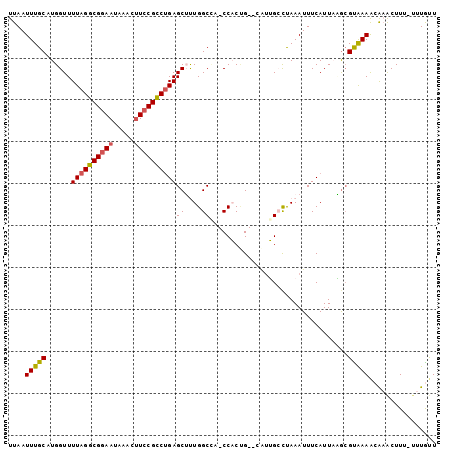
| Location | 15,872,888 – 15,872,986 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 79.61 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -15.51 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.32 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
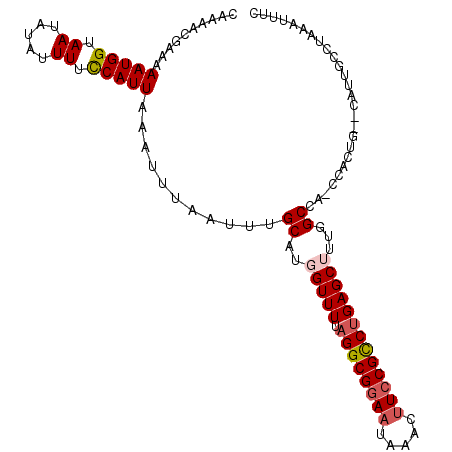
>2L_DroMel_CAF1 15872888 98 - 22407834 AACAAA-AAAGUUUGCUUUACGCCUAAUGAAAUUUAGGCAGUG--CGGUGG-UAGCCAAAGCUCAGGCGGAUGUGUAUUCCGCCUAAAACCAGGCAAAUUAA ......-..(((((((((...((((((......)))))).(.(--(..(((-...)))..)).)(((((((.......)))))))......))))))))).. ( -34.20) >DroPse_CAF1 38522 102 - 1 AAUAAGGAAAGCAUCUUUCACGCAUAAUGAAAUUUAACCCACAAACAGGGGCUGGCCAGCGCUCAGACGGAAGAUGAUUCCGCCUAAAACCAUGCAAAUUAA .....((((..((((((((..................(((.......))).(((((....)).)))..)))))))).))))..................... ( -22.60) >DroSec_CAF1 46363 98 - 1 AACAAA-AAAGUUUGCUUUGCGCUAAAUGAAAUUUAGGCAAUG--CAGUGG-CCGCCAAAGCUCAGGCGGAAGUUUAUUCCGCCUAAAACCAGGCAAAUUAA ......-.....((((.((((.((((((...)))))))))).)--)))..(-((((....))..((((((((.....)))))))).......)))....... ( -31.10) >DroSim_CAF1 46438 98 - 1 AACAAA-AAAGUUUGCUUUGCUUUGAAUGAAAUUUAGGCAAUG--CAGUGG-GCGCCAAAGCUCAGGCGGAAGUUUAUUUCGCCUAAAACCAGGCAAAUUAA ......-..((((((((((((.(((..(((...)))..))).)--))..((-((......))))((((((((.....))))))))......))))))))).. ( -28.30) >DroEre_CAF1 36421 97 - 1 AGCAA--AAAGUUCGUUUUACGCCUAAUGAAAUUUGGGCAAUA--CCGUGG-GAGCCAAAGCUCAGGCGGAAGCGUAUUCCGCCUAAAACCAUGCAAAUUAA .((..--..............((((((......))))))....--..((((-((((....))))((((((((.....))))))))....))))))....... ( -34.10) >DroPer_CAF1 64247 102 - 1 AAUAAGGAAAGCAUCUUUCACGCAUAAUGAAAUUUAACCCACAAACAGGGGCUGGCCAGCGCUCGGACGGAAGAUGAUUCCGCCUAAAACCAUGCAAAUUAA ......(((((...)))))..((((...((.......(((.......)))(((....)))..))((.(((((.....))))))).......))))....... ( -22.40) >consensus AACAAA_AAAGUUUGCUUUACGCAUAAUGAAAUUUAGGCAAUA__CAGUGG_UAGCCAAAGCUCAGGCGGAAGUUUAUUCCGCCUAAAACCAGGCAAAUUAA ..........((((.......((((((......))))))..........((...((....))..((((((((.....))))))))....))))))....... (-15.51 = -16.40 + 0.89)

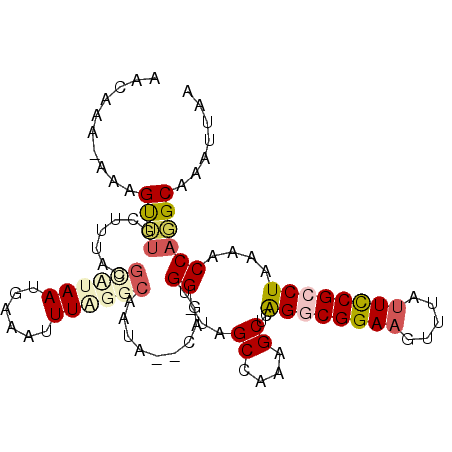
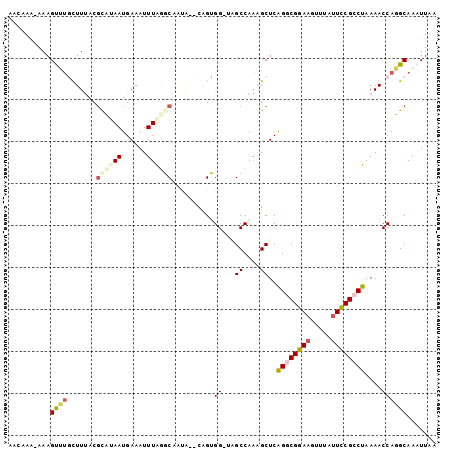
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:16 2006