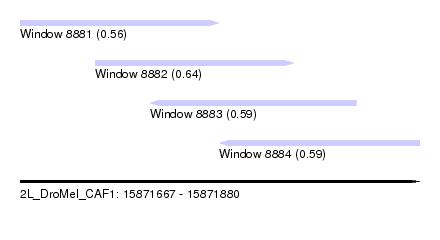
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,871,667 – 15,871,880 |
| Length | 213 |
| Max. P | 0.643361 |

| Location | 15,871,667 – 15,871,773 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -22.18 |
| Consensus MFE | -14.85 |
| Energy contribution | -15.55 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
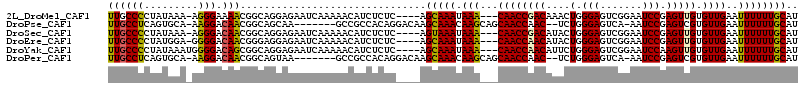
>2L_DroMel_CAF1 15871667 106 + 22407834 GCUUGUGAUUUCCCAAAUCCCAUUUAAGAAUUCCCUUUGUGGGAGCCAGGGAUAC------CCAGAACACAGGCU--------CACCCACAACAGUUGAACACACUCCUUUGCAUAAUUC (((((((.((((((...((((((..(((......))).))))))....))))...------...)).))))))).--------.........(((..((......))..)))........ ( -24.20) >DroPse_CAF1 37171 120 + 1 GUUUGUGAUUUCCCAAAUCCCAUUUAAGUAUUCCCUUUGUGGGAGACAGGGAUACGAUACUCCGGAACACACUCUCUCACGCGCACACCCAACAGUUGAACAAACUCUUUUGCAUAAUUC ((.(((((..((((...((((((..(((......))).))))))....))))..((......))............))))).))........(((..((......))..)))........ ( -22.90) >DroGri_CAF1 16697 87 + 1 GCUUGUGAUUUCCCAAAUCCCAUUUAAGUAUUCCCUUUAUGGGAC-----------------CACAACGA----------------ACCGCACAGUUCAACAAACUCUUUUGCAUAAUUC (((((((..........((((((..(((......))).)))))).-----------------))))).((----------------((......)))).............))....... ( -14.00) >DroEre_CAF1 35195 105 + 1 GCUUGUGAUUUCCCAAAUCCCAUUUAAGAAUUCCCUUUGUGGGAGCCUGGGACAC------CCAGAACUCAG-CC--------AGCCCACAACAGUUGAACCCACUCCUUUGCAUAAUUC ((..(((...(((((..((((((..(((......))).))))))...)))))...------.......((((-(.--------...........)))))...)))......))....... ( -23.20) >DroYak_CAF1 19818 106 + 1 GCUUGUGAUUUCCCAAAUCCCAUUUAAGAAUUCCCUUUGUGGGAGCCAGGGAUUC------CCGGAACACAGGCU--------CGCCCACAACAGUUGAACACACUCCUUUGCAUAAUUC (((((((..((((....((((((..(((......))).))))))....(((...)------))))))))))))).--------.........(((..((......))..)))........ ( -25.90) >DroPer_CAF1 62909 120 + 1 GUUUGUGAUUUCCCAAAUCCCAUUUAAGUAUUCCCUUUGUGGGAGACAGGGAUACGAUACUCCGGAACACACUCUCUCACGCGCACACCCAACAGUUGAACAAACUCUUUUGCAUAAUUC ((.(((((..((((...((((((..(((......))).))))))....))))..((......))............))))).))........(((..((......))..)))........ ( -22.90) >consensus GCUUGUGAUUUCCCAAAUCCCAUUUAAGAAUUCCCUUUGUGGGAGCCAGGGAUAC______CCAGAACACAG_CU________CACACACAACAGUUGAACAAACUCCUUUGCAUAAUUC ..(((((...((((...((((((..(((......))).))))))....))))........................................(((..((......))..))))))))... (-14.85 = -15.55 + 0.70)
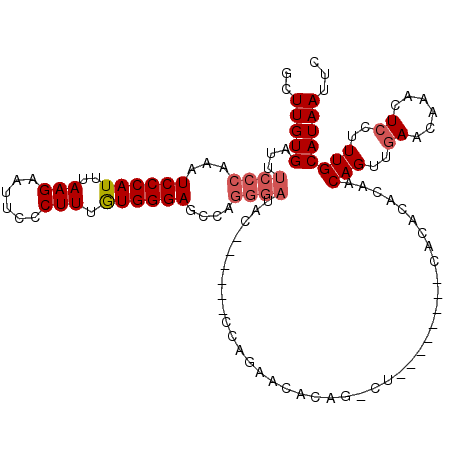

| Location | 15,871,707 – 15,871,813 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -19.34 |
| Energy contribution | -18.98 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15871707 106 + 22407834 GGGAGCCAGGGAUAC------CCAGAACACAGGCU--------CACCCACAACAGUUGAACACACUCCUUUGCAUAAUUCAUGCAAAAAAUUCAACACAACUCGGAUUCCGACUCCCAGU (((((...(((((.(------(.((......((..--------...))......((((((........(((((((.....)))))))...))))))....)).)))))))..)))))... ( -28.10) >DroPse_CAF1 37211 119 + 1 GGGAGACAGGGAUACGAUACUCCGGAACACACUCUCUCACGCGCACACCCAACAGUUGAACAAACUCUUUUGCAUAAUUCAUGCAAAAAAUUCAACACGACUCGGAUU-UGACUCCCAGA (((((.(((((...((.......(((......)))......))....)))....((((((.......((((((((.....))))))))..))))))............-)).)))))... ( -25.12) >DroSim_CAF1 45229 106 + 1 GGGAGCCAGGGAUAC------CCAGAACACAGGCU--------CGCCCACAACAGUUGAACACACUCCUUUGCAUAAUUCAUGCAAAAAAUUCAACACAACUCGGAUUCCGACUCCCAGU (((((...(((((.(------(.((......((..--------..)).......((((((........(((((((.....)))))))...))))))....)).)))))))..)))))... ( -28.90) >DroEre_CAF1 35235 105 + 1 GGGAGCCUGGGACAC------CCAGAACUCAG-CC--------AGCCCACAACAGUUGAACCCACUCCUUUGCAUAAUUCAUGCAAAAAAUUCAACACAACUCGGAUUCCGACUCCCAGU (((((.(((((...)------)))).......-..--------...........((((((........(((((((.....)))))))...)))))).....((((...)))))))))... ( -26.80) >DroYak_CAF1 19858 106 + 1 GGGAGCCAGGGAUUC------CCGGAACACAGGCU--------CGCCCACAACAGUUGAACACACUCCUUUGCAUAAUUCAUGCAAAAAAUUCAACACAACUUGGAUUCCGACUCCCAGA (((((.(.(((...)------))((((..((((..--------..)).......((((((........(((((((.....)))))))...))))))......))..))))).)))))... ( -29.50) >DroPer_CAF1 62949 119 + 1 GGGAGACAGGGAUACGAUACUCCGGAACACACUCUCUCACGCGCACACCCAACAGUUGAACAAACUCUUUUGCAUAAUUCAUGCAAAAAAUUCAACACGACUCGGAUU-UGACUCCCAGA (((((.(((((...((.......(((......)))......))....)))....((((((.......((((((((.....))))))))..))))))............-)).)))))... ( -25.12) >consensus GGGAGCCAGGGAUAC______CCAGAACACAGGCU________CACCCACAACAGUUGAACACACUCCUUUGCAUAAUUCAUGCAAAAAAUUCAACACAACUCGGAUUCCGACUCCCAGA (((((.................................................((((((........(((((((.....)))))))...)))))).....(((.....))))))))... (-19.34 = -18.98 + -0.36)

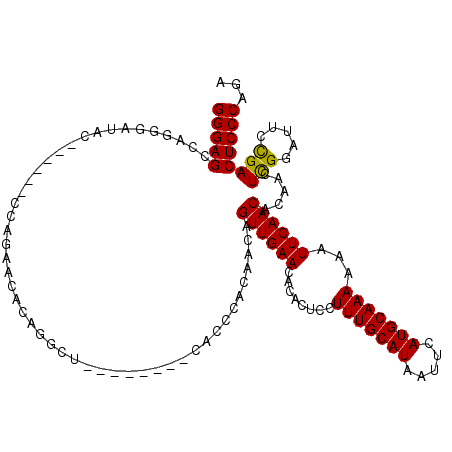
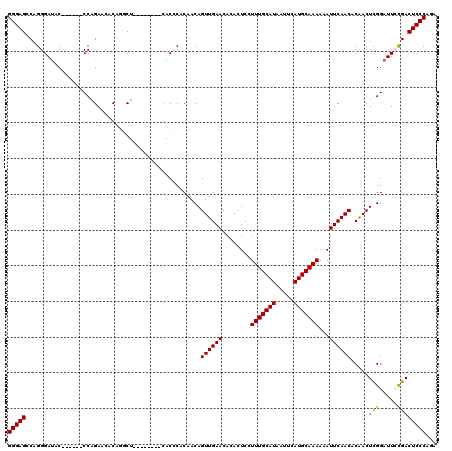
| Location | 15,871,736 – 15,871,846 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.87 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -20.39 |
| Energy contribution | -19.95 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15871736 110 - 22407834 AAAAACAUCUCUC----AGCAAAUAAA---CAACCGACAAACUGGGAGUCGGAAUCCGAGUUGUGUUGAAUUUUUUGCAUGAAUUAUGCAAAGGAGUGUGUUCAACUGUUGUGGGUG--- .....(((((..(----((((....((---((..(((((((((.(((.......))).)))).)))))..(((((((((((...)))))))))))...))))....))))).)))))--- ( -31.10) >DroPse_CAF1 37251 114 - 1 ---GCCGCCACAGGACAAGCAAACAAGCAGCAACCAAC--UCUGGGAGUCA-AAUCCGAGUCGUGUUGAAUUUUUUGCAUGAAUUAUGCAAAAGAGUUUGUUCAACUGUUGGGUGUGCGC ---((((((((((....(((((((...(((((....((--((..(((....-..)))))))..)))))..(((((((((((...))))))))))))))))))...))))..)))).)).. ( -38.40) >DroSec_CAF1 45197 110 - 1 AAAAACAUCUCUC----AGUAAAUAAA---CAACCGACAUACUGGGAGUCGGAAUCCGAGUUGUGUUGAAUUUUUUGCAUGAAUUAUGCAAAGGAGUGUGUUCAACUGUUGUGGGCG--- .....(((....(----(((.((((.(---(...(((((((((.(((.......))).)).)))))))..(((((((((((...))))))))))))).))))..))))..)))....--- ( -27.70) >DroEre_CAF1 35263 110 - 1 AAAAACAUCUCUC----AGCAAAUAAA---CAACCAACAUACUGGGAGUCGGAAUCCGAGUUGUGUUGAAUUUUUUGCAUGAAUUAUGCAAAGGAGUGGGUUCAACUGUUGUGGGCU--- ........(((.(----((((....((---(.(((((((((((.(((.......))).)).)))))))..(((((((((((...)))))))))))))..)))....))))).)))..--- ( -30.30) >DroYak_CAF1 19887 110 - 1 AAAAACAUCUCUC----AGCAAAUAAA---CAACCAACAUUCUGGGAGUCGGAAUCCAAGUUGUGUUGAAUUUUUUGCAUGAAUUAUGCAAAGGAGUGUGUUCAACUGUUGUGGGCG--- ........(((.(----((((.....(---((((....((((((.....))))))....)))))(((((((((((((((((...)))))))))).....)))))))))))).)))..--- ( -28.40) >DroPer_CAF1 62989 114 - 1 ---GCCGCCACAGGACAAGCAAACAAGCAGCAACCAAC--UCUGGGAGUCA-AAUCCGAGUCGUGUUGAAUUUUUUGCAUGAAUUAUGCAAAAGAGUUUGUUCAACUGUUGGGUGUGCGC ---((((((((((....(((((((...(((((....((--((..(((....-..)))))))..)))))..(((((((((((...))))))))))))))))))...))))..)))).)).. ( -38.40) >consensus AAAAACAUCUCUC____AGCAAAUAAA___CAACCAACAUACUGGGAGUCGGAAUCCGAGUUGUGUUGAAUUUUUUGCAUGAAUUAUGCAAAGGAGUGUGUUCAACUGUUGUGGGCG___ ..................................((((...((.(((.......))).))....(((((((((((((((((...)))))))))).....))))))).))))......... (-20.39 = -19.95 + -0.44)

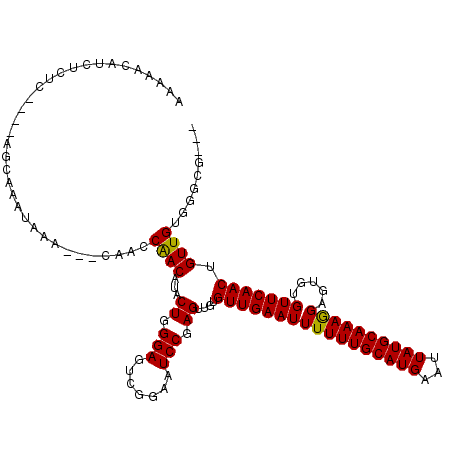
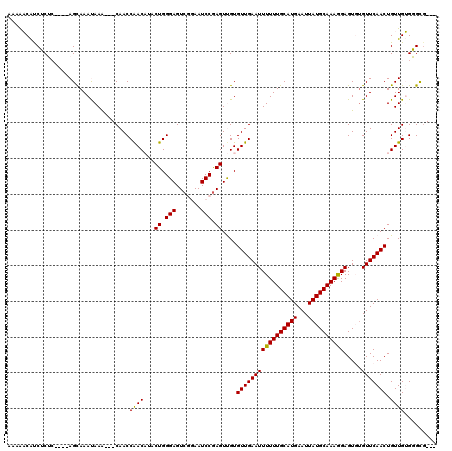
| Location | 15,871,773 – 15,871,880 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.89 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -15.10 |
| Energy contribution | -14.55 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15871773 107 - 22407834 UUGCCCCUAUAAA-AGGGAAAACGGCAGGAGAAUCAAAAACAUCUCUC----AGCAAAUAAA---CAACCGACAAACUGGGAGUCGGAAUCCGAGUUGUGUUGAAUUUUUUGCAU ....((((.....-))))......((((((((.((((.(((..(((((----((........---...........)))))))((((...)))))))...)))).)))))))).. ( -28.51) >DroPse_CAF1 37291 104 - 1 UUGCCUCAGUGCA-AAGGACAACGGCAGCAA-------GCCGCCACAGGACAAGCAAACAAGCAGCAACCAAC--UCUGGGAGUCA-AAUCCGAGUCGUGUUGAAUUUUUUGCAU ........(((((-(((((...((((.....-------))))(.(((.(((..((......))..........--....(((....-..)))..))).))).)..)))))))))) ( -26.50) >DroSec_CAF1 45234 107 - 1 UUGCCCCUAUAAA-AGGGACAACGGCAGGAGAAUCAAAAACAUCUCUC----AGUAAAUAAA---CAACCGACAUACUGGGAGUCGGAAUCCGAGUUGUGUUGAAUUUUUUGCAU (((.((((.....-)))).)))..((((((((.((((.(((..(((((----((((......---.........)))))))))((((...)))))))...)))).)))))))).. ( -32.66) >DroEre_CAF1 35300 107 - 1 UUGCCCCUAUGGA-GGGGACAACGGGAGGAGAAUCAAAAACAUCUCUC----AGCAAAUAAA---CAACCAACAUACUGGGAGUCGGAAUCCGAGUUGUGUUGAAUUUUUUGCAU ((((((((....)-)))).))).(((((..(.........)..)))))----.(((((....---....(((((((((.(((.......))).)).))))))).....))))).. ( -31.92) >DroYak_CAF1 19924 108 - 1 UUGCCCCUAUAAAUGGGGACAGCGGCAGGAGAAUCAAAAACAUCUCUC----AGCAAAUAAA---CAACCAACAUUCUGGGAGUCGGAAUCCAAGUUGUGUUGAAUUUUUUGCAU (((((((.......)))).)))..((((((((.((((.(((..(((((----((........---...........)))))))..((...))..)))...)))).)))))))).. ( -27.91) >DroPer_CAF1 63029 104 - 1 UUGCCUCAGUGCA-AAGGACAACGGCAGUAA-------GCCGCCACAGGACAAGCAAACAAGCAGCAACCAAC--UCUGGGAGUCA-AAUCCGAGUCGUGUUGAAUUUUUUGCAU ........(((((-(((((...((((.....-------))))(.(((.(((..((......))..........--....(((....-..)))..))).))).)..)))))))))) ( -26.50) >consensus UUGCCCCUAUAAA_AGGGACAACGGCAGGAGAAUCAAAAACAUCUCUC____AGCAAAUAAA___CAACCAACAUACUGGGAGUCGGAAUCCGAGUUGUGUUGAAUUUUUUGCAU ((((((.........))).)))...............................(((((.(((...((((((((....(.(((.......))).))))).))))..)))))))).. (-15.10 = -14.55 + -0.55)
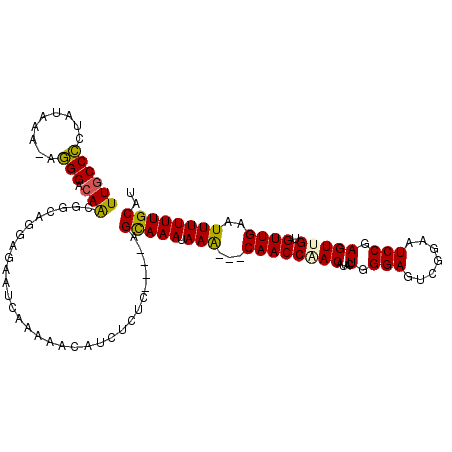
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:10 2006