| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,855,573 – 15,855,782 |
| Length | 209 |
| Max. P | 0.904021 |

| Location | 15,855,573 – 15,855,667 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -32.21 |
| Consensus MFE | -23.11 |
| Energy contribution | -23.44 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15855573 94 - 22407834 ---------CAUUCAACCCAUUUACAUGCAA----------------CGCAGGCAG-GGCGGAAACGGAAGUGCCGAGGCAGCUCUGAAAUAAUAUCAAAAAUGGCUGCCGGAGGCGGCA ---------..(((..(((.......(((..----------------.)))....)-))..)))((....))((((.(((((((.(((.......))).....))))))).....)))). ( -28.50) >DroPse_CAF1 17649 105 - 1 ---------------CUCCAUUUACAUGCAGUCCUGCAAGUCGACGGCGCAGGCCGAAGCGGAAACGGAAGUGCAGAGGCAGCUCAGAAAUAAUAUCAAAAAUGGCUGCCGGAGGCGGCA ---------------((((..............(((((..((..((((....))))...((....))))..))))).((((((.((................))))))))))))...... ( -39.09) >DroSec_CAF1 27830 94 - 1 ---------CAUUCAACCCAUUUACAUGCAA----------------CGCAGGCAG-GGCGGAAACGGAAGUGCCGAGGCAGCUCUGAAAUAAUAUCAAAAAUGGCUGCCGGAGGCGGCA ---------..(((..(((.......(((..----------------.)))....)-))..)))((....))((((.(((((((.(((.......))).....))))))).....)))). ( -28.50) >DroSim_CAF1 28864 94 - 1 ---------CAUUCAACCCAUUUACAUGCAA----------------CGCAGGCAG-GGCGGAAACGGAAGUGCCGAGGCAGCUCUGAAAUAAUAUCAAAAAUGGCUGCCGGAGGCGGCA ---------..(((..(((.......(((..----------------.)))....)-))..)))((....))((((.(((((((.(((.......))).....))))))).....)))). ( -28.50) >DroEre_CAF1 15194 103 - 1 CAUGCAACCCAUUCAACCCAUUUACAUGCAA----------------CGCGGGCAG-GGCGGAAACGGAAGUGCCGAGGCAGCUCUGAAAUAAUAUCAAAAAUGGCUGCCGGAGGCGGCA ..(((......(((..(((.......(((..----------------.....))))-))..)))((....))(((..(((((((.(((.......))).....)))))))...))).))) ( -29.61) >DroPer_CAF1 43410 105 - 1 ---------------CUCCAUUUACAUGCAGUCCUGCAAGUCGACGGCGCAGGCCGAAGCGGAAACGGAAGUGCAGAGGCAGCUCAGAAAUAAUAUCAAAAAUGGCUGCCGGAGGCGGCA ---------------((((..............(((((..((..((((....))))...((....))))..))))).((((((.((................))))))))))))...... ( -39.09) >consensus _________CAUUCAACCCAUUUACAUGCAA________________CGCAGGCAG_GGCGGAAACGGAAGUGCCGAGGCAGCUCUGAAAUAAUAUCAAAAAUGGCUGCCGGAGGCGGCA ..........................(((..................(((........)))...((....))(((..(((((((...................)))))))...))).))) (-23.11 = -23.44 + 0.33)

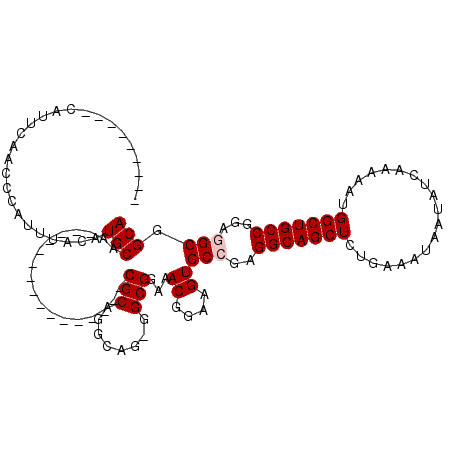

| Location | 15,855,645 – 15,855,742 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 89.90 |
| Mean single sequence MFE | -20.76 |
| Consensus MFE | -19.26 |
| Energy contribution | -19.57 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.904021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15855645 97 - 22407834 GCACCAUUGCAUUCUGAAGUUCUUGCACUCUGCACUCCCUUUAGCCCCGAUAAGCGAUCGCAAA-AAAGGGACUCC---------CAUUCAACCCAUUUACAUGCAA ......((((((..((((.....(((.....))).(((((((.....((((.....))))....-)))))))....---------...........)))).)))))) ( -17.40) >DroSec_CAF1 27902 98 - 1 GCACCAUUGCAUUCUGAAGCUCUUGCACGCUGCACUCCCUUUAGCCCCGAUAAGCGAUCGCAAAAAAAGGGAGUCU---------CAUUCAACCCAUUUACAUGCAA ......((((((..((((((....)).......(((((((((.....((((.....)))).....)))))))))..---------...........)))).)))))) ( -21.70) >DroSim_CAF1 28936 97 - 1 GCACCAUUGCAUUCUAAAGCUCUUGCACUCUGCACUCCCUUUAGCCCCGAUAAGCGAUCGCAAA-AAAGGGAGUCU---------CAUUCAACCCAUUUACAUGCAA ......((((((..((((((....)).......(((((((((.....((((.....))))....-)))))))))..---------...........)))).)))))) ( -21.20) >DroEre_CAF1 15266 107 - 1 GCACCAUUGCAUUCUGAAGCUCCUGCACUCGGCACUCCCUUUGGCCCCGAUAAGCGAUCUCAAAAAAAGGGAGCCCCAUGCAACCCAUUCAACCCAUUUACAUGCAA (((...........((((.....((((...((..((((((((......(((.....)))......))))))))..)).)))).....))))...........))).. ( -22.75) >consensus GCACCAUUGCAUUCUGAAGCUCUUGCACUCUGCACUCCCUUUAGCCCCGAUAAGCGAUCGCAAA_AAAGGGAGUCC_________CAUUCAACCCAUUUACAUGCAA ......((((((..((((.....(((.....)))((((((((.....((((.....)))).....)))))))).......................)))).)))))) (-19.26 = -19.57 + 0.31)

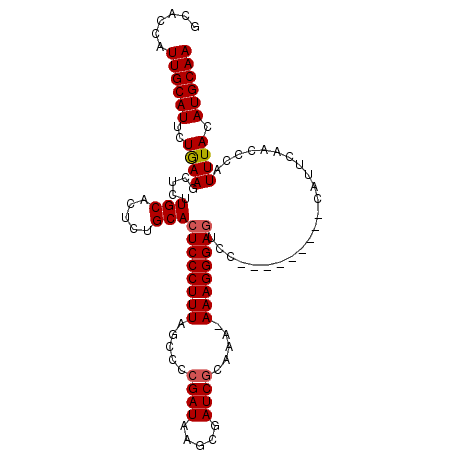

| Location | 15,855,667 – 15,855,782 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 87.63 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -21.46 |
| Energy contribution | -21.40 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15855667 115 - 22407834 UCAGUCGCCCACAUUAACUUCAAUUCGCUCCCUAGCACACGCACCAUUGCAUUCUGAAGUUCUUGCACUCUGCACUCCCUUUAGCCCCGAUAAGCGAUCGCAAA-AAAGGGACUCC ......((...((..(((((((....(((....)))....(((....)))....)))))))..))......))..(((((((.....((((.....))))....-))))))).... ( -22.90) >DroSec_CAF1 27924 100 - 1 ----------------ACUUCAAUUCGCUCCCUAGCACACGCACCAUUGCAUUCUGAAGCUCUUGCACGCUGCACUCCCUUUAGCCCCGAUAAGCGAUCGCAAAAAAAGGGAGUCU ----------------.(((((....(((....)))....(((....)))....))))).....((.....))(((((((((.....((((.....)))).....))))))))).. ( -24.30) >DroSim_CAF1 28958 115 - 1 UCAGUCGCCCACAUUAACUUCAAUUCGCUCCCUAGCACACGCACCAUUGCAUUCUAAAGCUCUUGCACUCUGCACUCCCUUUAGCCCCGAUAAGCGAUCGCAAA-AAAGGGAGUCU ..........................(((((((.((....))....((((...((((((....(((.....)))....))))))....(((.....))))))).-..))))))).. ( -24.10) >DroEre_CAF1 15297 116 - 1 UCAGUCGCCCACAUUAACUUCAAUUCGCUCCCUCGCACACGCACCAUUGCAUUCUGAAGCUCCUGCACUCGGCACUCCCUUUGGCCCCGAUAAGCGAUCUCAAAAAAAGGGAGCCC ..........................(((((((.((....))...(((((........((....))..((((..((......))..))))...))))).........))))))).. ( -24.40) >consensus UCAGUCGCCCACAUUAACUUCAAUUCGCUCCCUAGCACACGCACCAUUGCAUUCUGAAGCUCUUGCACUCUGCACUCCCUUUAGCCCCGAUAAGCGAUCGCAAA_AAAGGGAGUCC ..........................(((((((.((....))....((((...((((((....(((.....)))....))))))....(((.....)))))))....))))))).. (-21.46 = -21.40 + -0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:57 2006