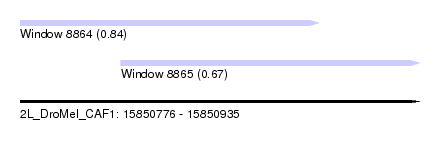
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,850,776 – 15,850,935 |
| Length | 159 |
| Max. P | 0.839731 |

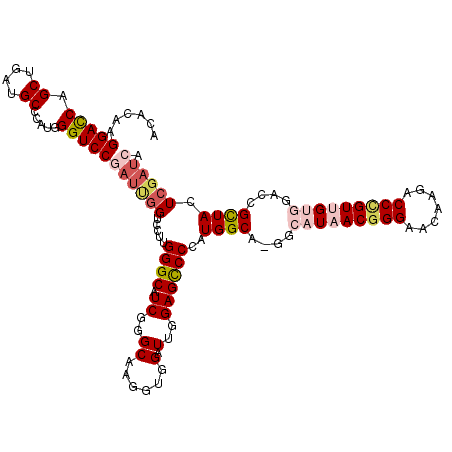
| Location | 15,850,776 – 15,850,895 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.21 |
| Mean single sequence MFE | -47.83 |
| Consensus MFE | -18.88 |
| Energy contribution | -21.48 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15850776 119 + 22407834 AUCCACCGUGCUACUUGCCCCAACAGGGCGAUGAGUUGCCUCACAAGGACCAGCUGAUGCCCAUGGGUCCGAUUGGUCCAUGGGCAUCGGGCAAGGUGGAUUGGAGCCCCAUGGCC-GGC (((((((.((((..((((((.....))))))..((((((((....)))..)))))(((((((((((..(.....)..))))))))))).)))).)))))))..(.(((....))))-... ( -57.80) >DroPse_CAF1 12713 119 + 1 AUCCGCCAUGCUAUUUGCCGCAGGCGGGCGACGAUCUGCCACACAAGGAUCAGCUGAUGCCAAUGGGUCCGAUCGGGCCCUGGGCUUCUGGCAAGGUCGAUUGGAGUCCUAUGGCA-GGC ....(((.((((((..(((...)))((((..(((((.(((.(....).....(((((.(((...((((((....))))))..))).)).)))..))).)))))..)))).))))))-))) ( -52.10) >DroSec_CAF1 23021 119 + 1 AUCCACCGUGCUACUUGCCCCAACAGGGCGAUGAGCUGCCUCACAAGGACCAGCUGAUGCCCAUGGGUCCGAUUGGUCCUUGGGCAUCGGGCAAGGUGGAUUGGAGCCCCAUGGCC-GGC (((((((.((((..((((((.....))))))..((((((((....)))..)))))((((((((.(((.((....)).))))))))))).)))).)))))))..(.(((....))))-... ( -56.10) >DroSim_CAF1 23385 120 + 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGUCCUUGGGCAUCGGGCAAGGGGGAUUGGAGCCCCCUGGCAGGGC ...........................................................................(((....))).((..((.((((((.......)))))).))..)). ( -17.80) >DroEre_CAF1 10506 119 + 1 AUCCACCGUGCUACUUGCCCCAACAGGGGGAUGAGUUGCCCCACAAGGACCAGCUAAUGCCCAUGGGUCCAAUUGGUCCUUGGGCAUCGGGCAAAGUAGAUUGGAGCCCCAUGGCC-GGG ..((.(((..((((((((((.....((((.(.....).)))).((((((((((.....(((....)))....))))))))))......)))).))))))..))).(((....))).-)). ( -51.10) >DroPer_CAF1 38476 119 + 1 AUCCGCCAUGCUAUUUGCCGCAGGCGGGCGACGAUCUGCCACACAAGGAUCAGCUGAUGCCAAUGGGUCCGAUCGGGCCCUGGGCUUCUGGCAAGGUCGAUUGGAGUCCUAUGGCA-GGC ....(((.((((((..(((...)))((((..(((((.(((.(....).....(((((.(((...((((((....))))))..))).)).)))..))).)))))..)))).))))))-))) ( -52.10) >consensus AUCCACCGUGCUACUUGCCCCAACAGGGCGAUGAGCUGCCACACAAGGACCAGCUGAUGCCCAUGGGUCCGAUUGGUCCUUGGGCAUCGGGCAAGGUGGAUUGGAGCCCCAUGGCA_GGC .....(((.....(((((((......((((......))))..................(((...(((.((....)).)))..)))...)))))))......))).(((....)))..... (-18.88 = -21.48 + 2.60)

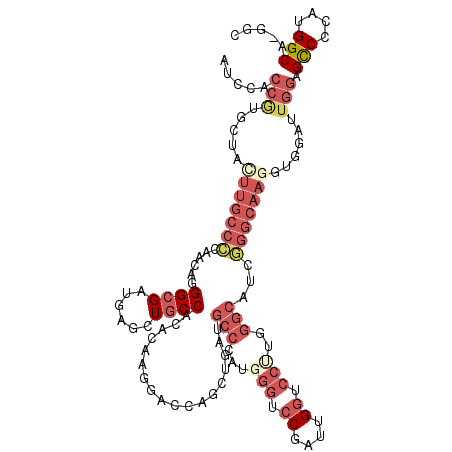
| Location | 15,850,816 – 15,850,935 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -46.36 |
| Consensus MFE | -25.87 |
| Energy contribution | -27.38 |
| Covariance contribution | 1.51 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15850816 119 + 22407834 UCACAAGGACCAGCUGAUGCCCAUGGGUCCGAUUGGUCCAUGGGCAUCGGGCAAGGUGGAUUGGAGCCCCAUGGCC-GGCAUAACGGGAACGAGACCCGUUGUGGACCGCUACUCGAUCA ........(((.((((((((((((((..(.....)..))))))))))).)))..))).((((((.(((....)))(-(((((((((((.......)))))))))..)))....)))))). ( -53.80) >DroPse_CAF1 12753 119 + 1 ACACAAGGAUCAGCUGAUGCCAAUGGGUCCGAUCGGGCCCUGGGCUUCUGGCAAGGUCGAUUGGAGUCCUAUGGCA-GGCCUGACGGGAACACGUCCUGUGGUAGAUCGUUACUCGAUCA .((((.(((((((((..(((((..((((((....)))))).(((((((((((...))))...)))))))..)))))-)).))))((......)))))))))...(((((.....))))). ( -47.10) >DroSec_CAF1 23061 119 + 1 UCACAAGGACCAGCUGAUGCCCAUGGGUCCGAUUGGUCCUUGGGCAUCGGGCAAGGUGGAUUGGAGCCCCAUGGCC-GGCAUAACGGGAACGAGACCCGUUGUGGACCGCUACUCGAUCA ........(((.(((((((((((.(((.((....)).))))))))))).)))..))).((((((.(((....)))(-(((((((((((.......)))))))))..)))....)))))). ( -49.70) >DroSim_CAF1 23425 120 + 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGUCCUUGGGCAUCGGGCAAGGGGGAUUGGAGCCCCCUGGCAGGGCAUAACGGGAACGGGCCCCGUUGUGGACCGCUACUCGAUCA ...................................(((....)))((((((..((((((.......))))))(((.((.(((((((((.......)))))))))..))))).)))))).. ( -39.10) >DroEre_CAF1 10546 119 + 1 CCACAAGGACCAGCUAAUGCCCAUGGGUCCAAUUGGUCCUUGGGCAUCGGGCAAAGUAGAUUGGAGCCCCAUGGCC-GGGAUAACAGGCACAAGACCCGUUGUGGACCGCUACUCGAUCA ((....))....(((.(((((((.(((.((....)).))))))))))..)))......(((((((((.(((..((.-(((...............)))))..)))...))).).))))). ( -41.06) >DroPer_CAF1 38516 119 + 1 ACACAAGGAUCAGCUGAUGCCAAUGGGUCCGAUCGGGCCCUGGGCUUCUGGCAAGGUCGAUUGGAGUCCUAUGGCA-GGCCUGACGGGAACACGUCCUGUGGUGGAUCGUUACUCGAUCA .(((.((((((((((..(((((..((((((....)))))).(((((((((((...))))...)))))))..)))))-)).))))((......))))))...)))(((((.....))))). ( -47.40) >consensus ACACAAGGACCAGCUGAUGCCCAUGGGUCCGAUUGGUCCUUGGGCAUCGGGCAAGGUGGAUUGGAGCCCCAUGGCA_GGCAUAACGGGAACAAGACCCGUUGUGGACCGCUACUCGAUCA ......(((((.((....)).....)))))((((((.....((((.((..((......).)..))))))..((((....(((((((((.......)))))))))....)))).)))))). (-25.87 = -27.38 + 1.51)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:51 2006