
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,848,171 – 15,848,370 |
| Length | 199 |
| Max. P | 0.968689 |

| Location | 15,848,171 – 15,848,279 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 97.69 |
| Mean single sequence MFE | -39.75 |
| Consensus MFE | -37.51 |
| Energy contribution | -37.58 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
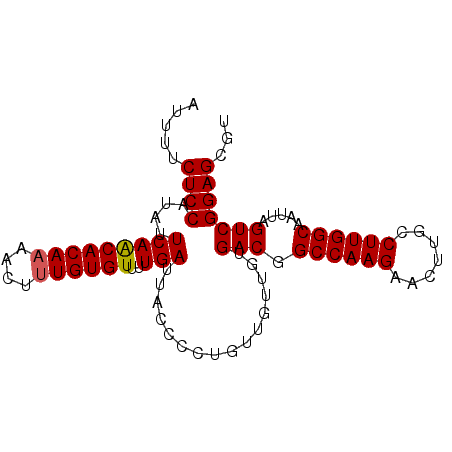
>2L_DroMel_CAF1 15848171 108 - 22407834 CUAAGUAACGCUCUCUGGGAACUUCAUGAAGUGAGCAACUUUGGAGUGGCCCCGGGGUGCAAUGAUGCCAUCAAAAUUGGCGCUGCAACAAGCGGCGGAACAAAGAGU ....((..(((.(((((((.((((((..((((.....))))))))))...))))))).))......((((.......))))(((((.....))))))..))....... ( -38.40) >DroSec_CAF1 20429 108 - 1 CUAAGUAACGCUCUCCGGGAACUUCUUGAAGUGAGCAACUUUGGAGUGGCCCCGGGGUGCAAUGAUGCCAUCAAAAUUGGCGCUGCAACAAGCGGCGGAACAAAGAGU ....((..(((.(((((((.(((((..(((((.....))))))))))...))))))).))......((((.......))))(((((.....))))))..))....... ( -40.30) >DroSim_CAF1 20823 108 - 1 CUAAGUAACGCUCUCCGGGAACUUCUUGAAGUGAGCAACUUUGGAGUGGCCCCGGGGUGCAAUGAUGCCAUCAAAAUUGGCGCUGCAACAAGCGGCGGAACAAAGAGU ....((..(((.(((((((.(((((..(((((.....))))))))))...))))))).))......((((.......))))(((((.....))))))..))....... ( -40.30) >DroEre_CAF1 5394 108 - 1 CUAAGUAACGCUCUCCGGGAACUUCUUGAAGUGAGCAACUUUGGAGUGGCCCCGGGGUGCAAUGAUGGCAUCAAAAUUGGCGCUGCAACAAGCGGCGAAACAAAGAGC .........((((((((((.(((((..(((((.....))))))))))...)))))(((((.......)))))....(((.((((((.....))))))...)))))))) ( -40.00) >consensus CUAAGUAACGCUCUCCGGGAACUUCUUGAAGUGAGCAACUUUGGAGUGGCCCCGGGGUGCAAUGAUGCCAUCAAAAUUGGCGCUGCAACAAGCGGCGGAACAAAGAGU ....((..(((.(((((((.(((((..(((((.....))))))))))...))))))).))......((((.......))))(((((.....))))))..))....... (-37.51 = -37.58 + 0.06)

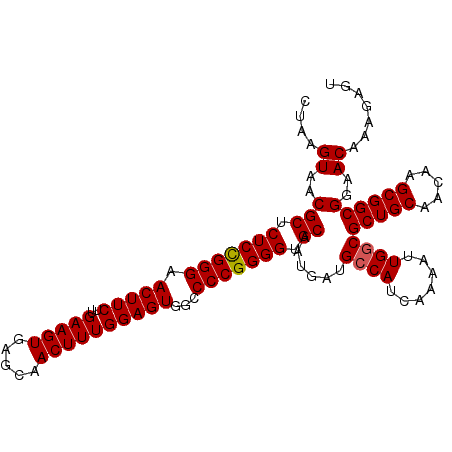
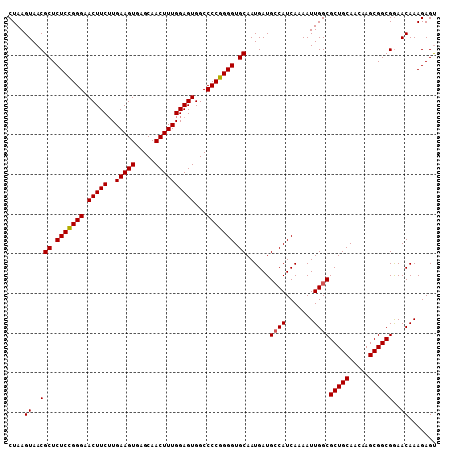
| Location | 15,848,211 – 15,848,315 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 97.60 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -31.60 |
| Energy contribution | -32.10 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15848211 104 + 22407834 GCAUCAUUGCACCCCGGGGCCACUCCAAAGUUGCUCACUUCAUGAAGUUCCCAGAGAGCGUUACUUAGACGCUCCGACUAAUUGCCAAGGCAAGUUCUUGGCCG (((....))).......(((((......(((((...((((....)))).......(((((((.....))))))))))))..((((....)))).....))))). ( -30.40) >DroSec_CAF1 20469 103 + 1 GCAUCAUUGCACCCCGGGGCCACUCCAAAGUUGCUCACUUCAAGAAGUUCCCGGAGAGCGUUACUUAGACGCUCCGACUAAUUGCCAAGGU-AGUUCUUGGCCG (((....)))...(((((((.(((....))).))..((((....)))).))))).(((((((.....))))))).........(((((((.-...))))))).. ( -35.60) >DroSim_CAF1 20863 104 + 1 GCAUCAUUGCACCCCGGGGCCACUCCAAAGUUGCUCACUUCAAGAAGUUCCCGGAGAGCGUUACUUAGACGCUCCGACUAAUUGCCAAGGCAAGUUCUUGGCCG (((....)))...(((((((.(((....))).))..((((....)))).))))).(((((((.....))))))).........(((((((.....))))))).. ( -34.70) >DroEre_CAF1 5434 104 + 1 CCAUCAUUGCACCCCGGGGCCACUCCAAAGUUGCUCACUUCAAGAAGUUCCCGGAGAGCGUUACUUAGACGCUCCGACUAAUUGCCAAGGCAAGUUCUUGGCCG ...((........(((((((.(((....))).))..((((....)))).))))).(((((((.....))))))).))......(((((((.....))))))).. ( -32.10) >consensus GCAUCAUUGCACCCCGGGGCCACUCCAAAGUUGCUCACUUCAAGAAGUUCCCGGAGAGCGUUACUUAGACGCUCCGACUAAUUGCCAAGGCAAGUUCUUGGCCG (((....)))...(((((((.(((....))).))..((((....)))).))))).(((((((.....))))))).........(((((((.....))))))).. (-31.60 = -32.10 + 0.50)

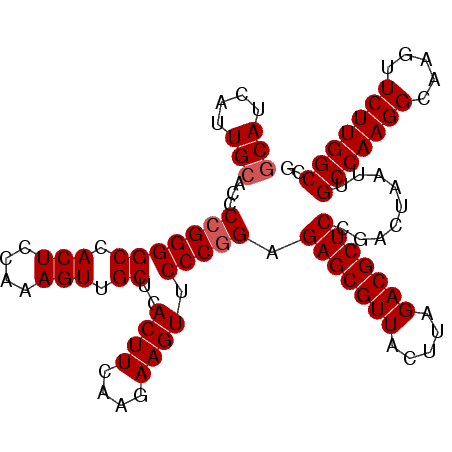
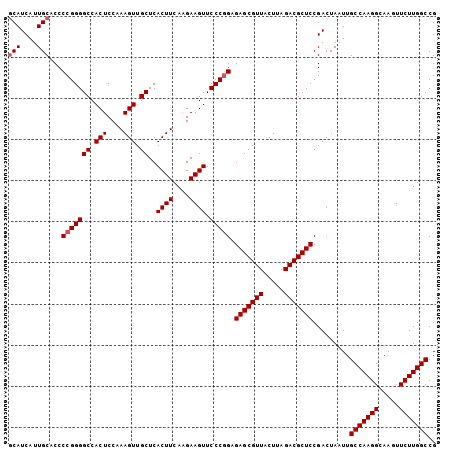
| Location | 15,848,246 – 15,848,355 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 95.41 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -31.88 |
| Energy contribution | -31.75 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15848246 109 + 22407834 CACUUCAUGAAGUUCCCAGAGAGCGUUACUUAGACGCUCCGACUAAUUGCCAAGGCAAGUUCUUGGCCGUCACAACAACAGGGGUAAUCAAGACACAAAGUUUUGUGUU .......(((...((((.(.(((((((.....))))))))(((.....(((((((.....))))))).))).........))))...))).(((((((....))))))) ( -33.70) >DroSec_CAF1 20504 108 + 1 CACUUCAAGAAGUUCCCGGAGAGCGUUACUUAGACGCUCCGACUAAUUGCCAAGGU-AGUUCUUGGCCGUCACAACAACAGGGGUGAUCAAAACACAAAGUUUUGUGUU ........((...((((.(.(((((((.....))))))))(((.....(((((((.-...))))))).))).........))))...))..(((((((....))))))) ( -33.20) >DroSim_CAF1 20898 109 + 1 CACUUCAAGAAGUUCCCGGAGAGCGUUACUUAGACGCUCCGACUAAUUGCCAAGGCAAGUUCUUGGCCGUCACAACAACAGGGGUAAUCAAAACACAAAGUUUUGUGUU ........((...((((.(.(((((((.....))))))))(((.....(((((((.....))))))).))).........))))...))..(((((((....))))))) ( -32.30) >DroEre_CAF1 5469 109 + 1 CACUUCAAGAAGUUCCCGGAGAGCGUUACUUAGACGCUCCGACUAAUUGCCAAGGCAAGUUCUUGGCCGUCACAAGAACAGGGGUAAUCAAAACACAGCGUUUUGUGCU .((((....)))).(((.(.(((((((.....))))))))......((((....))))(((((((.......))))))).)))(((..((((((.....))))))))). ( -33.10) >consensus CACUUCAAGAAGUUCCCGGAGAGCGUUACUUAGACGCUCCGACUAAUUGCCAAGGCAAGUUCUUGGCCGUCACAACAACAGGGGUAAUCAAAACACAAAGUUUUGUGUU ........((...((((.(.(((((((.....))))))))(((.....(((((((.....))))))).))).........))))...))..(((((((....))))))) (-31.88 = -31.75 + -0.12)

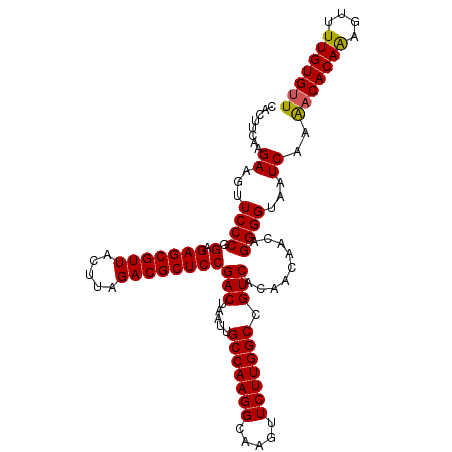
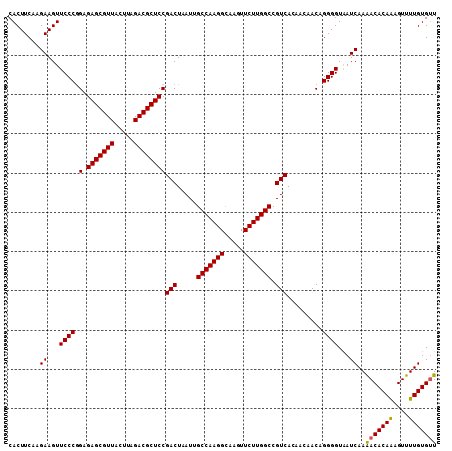
| Location | 15,848,279 – 15,848,370 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 94.51 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -22.83 |
| Energy contribution | -22.70 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15848279 91 + 22407834 ACGCUCCGACUAAUUGCCAAGGCAAGUUCUUGGCCGUCACAACAACAGGGGUAAUCAAGACACAAAGUUUUGUGUUGAAUAUGGAGAAAUG ...(((((((.....(((((((.....))))))).))).(((((((....))...((((((.....))))))))))).....))))..... ( -23.70) >DroSec_CAF1 20537 90 + 1 ACGCUCCGACUAAUUGCCAAGGU-AGUUCUUGGCCGUCACAACAACAGGGGUGAUCAAAACACAAAGUUUUGUGUUGAAUAUGGAGAAAAU ...(((((.......(((((((.-...))))))).(((((..(....)..)))))...(((((((....))))))).....)))))..... ( -26.80) >DroSim_CAF1 20931 91 + 1 ACGCUCCGACUAAUUGCCAAGGCAAGUUCUUGGCCGUCACAACAACAGGGGUAAUCAAAACACAAAGUUUUGUGUUGAAUAUGGAGAAAAU ...(((((((.....(((((((.....))))))).))).(((((((....))...((((((.....))))))))))).....))))..... ( -23.60) >DroEre_CAF1 5502 91 + 1 ACGCUCCGACUAAUUGCCAAGGCAAGUUCUUGGCCGUCACAAGAACAGGGGUAAUCAAAACACAGCGUUUUGUGCUGAAUAUGGAGAAAAU ...((((((((..((((....))))(((((((.......)))))))...)))..........(((((.....)))))....)))))..... ( -26.10) >consensus ACGCUCCGACUAAUUGCCAAGGCAAGUUCUUGGCCGUCACAACAACAGGGGUAAUCAAAACACAAAGUUUUGUGUUGAAUAUGGAGAAAAU ...(((((((.....(((((((.....))))))).)))............(((.(((..((((((....))))))))).)))))))..... (-22.83 = -22.70 + -0.13)

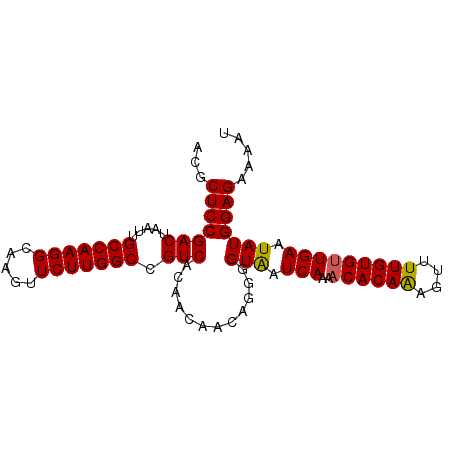
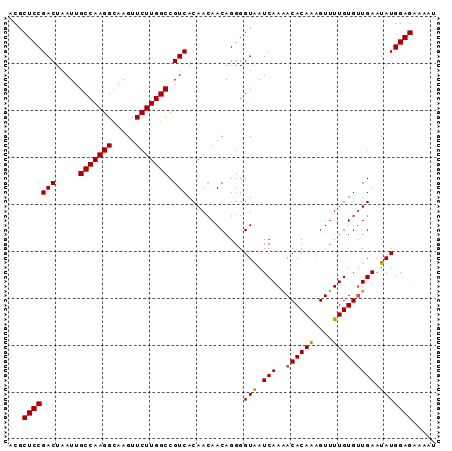
| Location | 15,848,279 – 15,848,370 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 94.51 |
| Mean single sequence MFE | -23.88 |
| Consensus MFE | -22.51 |
| Energy contribution | -22.58 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15848279 91 - 22407834 CAUUUCUCCAUAUUCAACACAAAACUUUGUGUCUUGAUUACCCCUGUUGUUGUGACGGCCAAGAACUUGCCUUGGCAAUUAGUCGGAGCGU .....((((.((.(((((((((....))))))..))).)).............(((.((((((.......)))))).....)))))))... ( -22.90) >DroSec_CAF1 20537 90 - 1 AUUUUCUCCAUAUUCAACACAAAACUUUGUGUUUUGAUCACCCCUGUUGUUGUGACGGCCAAGAACU-ACCUUGGCAAUUAGUCGGAGCGU .....(((((((..(((((((((((.....))))))........))))).)))(((.((((((....-..)))))).....)))))))... ( -24.50) >DroSim_CAF1 20931 91 - 1 AUUUUCUCCAUAUUCAACACAAAACUUUGUGUUUUGAUUACCCCUGUUGUUGUGACGGCCAAGAACUUGCCUUGGCAAUUAGUCGGAGCGU .....(((((((..(((((((((((.....))))))........))))).)))(((.((((((.......)))))).....)))))))... ( -24.00) >DroEre_CAF1 5502 91 - 1 AUUUUCUCCAUAUUCAGCACAAAACGCUGUGUUUUGAUUACCCCUGUUCUUGUGACGGCCAAGAACUUGCCUUGGCAAUUAGUCGGAGCGU .....((((......((((((......))))))..(((((.....(((((((.(....))))))))((((....)))).)))))))))... ( -24.10) >consensus AUUUUCUCCAUAUUCAACACAAAACUUUGUGUUUUGAUUACCCCUGUUGUUGUGACGGCCAAGAACUUGCCUUGGCAAUUAGUCGGAGCGU .....((((....(((((((((....))))))..)))................(((.((((((.......)))))).....)))))))... (-22.51 = -22.58 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:50 2006