| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,844,230 – 15,844,337 |
| Length | 107 |
| Max. P | 0.543502 |

| Location | 15,844,230 – 15,844,337 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -18.97 |
| Energy contribution | -18.50 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
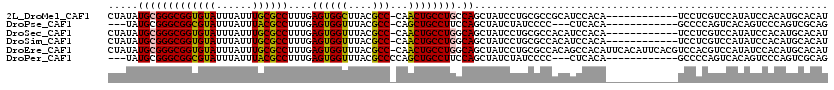
>2L_DroMel_CAF1 15844230 107 + 22407834 CUAUAUGCGGGCGGUGUAUUUAUUUGCGCCUUUGAGUGGCUUACGCC-CAACUGCCUGCCAGCUAUCCUGCGCCGCAUCCACA------------UCCUCGUCCAUAUCCACAUGCACAU ....((((((((((((((......))))))....((((((....((.-.....))..)))).)).....)).)))))).....------------.....((.(((......))).)).. ( -27.60) >DroPse_CAF1 3743 101 + 1 ---UAUGCGGGCGGCGUAUUUAUUUACGCCUUUGAGUGGUUUACGCC-CAGCUGCCUUCCAGCUAUCUAUCCCC---CUCACA------------GCCCCAGUCACAGUCCCAGUCGCAG ---..((.((((((((((......))))))..((((.((..((.(..-.(((((.....)))))..)))..)).---))))..------------))))))((.((.......)).)).. ( -31.00) >DroSec_CAF1 16456 107 + 1 CUAUAUGCGGGCGGUGUAUUUAUUUGCGCCUUUGAGUGGUUUACGCC-CAACUGCCUGGCAGCUAUCCUGCGCCACAUCCACA------------UCCUCGUCCAUAUCCACAUGCACAU .....(((((((((((((......)))))).(((.(((.....))).-)))..))))(((.((......))))).........------------...................)))... ( -27.80) >DroSim_CAF1 16764 107 + 1 CUAUAUGCGGGCGGUGUAUUUAUUUGCGCCUUUGAGUGGUUUACGCC-CAACUGCCUGGCAGCUAUCCUGCGCCACAUCCACA------------UCCUCGUCCAUAUCCACAUGCACAU .....(((((((((((((......)))))).(((.(((.....))).-)))..))))(((.((......))))).........------------...................)))... ( -27.80) >DroEre_CAF1 1339 119 + 1 CUAUAUGCGGGCGGUGUAUUUAUUUGCGCCUUUGAGUGGUUUACGCC-CAACUGCCUGGCAGCUAUCCUGCGCCACAGCCACAUUCACAUUCACGUCCACGUCCAUAUCCACAUGCACAU .....(((((((((((((......)))))).....((((((...((.-.....)).((((.((......)))))).))))))............))))..((........))..)))... ( -31.50) >DroPer_CAF1 29534 102 + 1 ---UAUGCGGGCGGCGUAUUUAUUUACGCCUUUGAGUGGUUUACGCCCCAGCUGCCUUCCAGCUAUCUAUCCCC---CUCACA------------GCCCCAGUCACAGUCCCAGUCGCAG ---..((.((((((((((......))))))..((((.((..((......(((((.....)))))...))..)).---))))..------------))))))((.((.......)).)).. ( -30.40) >consensus CUAUAUGCGGGCGGUGUAUUUAUUUGCGCCUUUGAGUGGUUUACGCC_CAACUGCCUGCCAGCUAUCCUGCGCCACAUCCACA____________UCCUCGUCCAUAUCCACAUGCACAU .....(((((((((((((......))))))....((((((....)))...)))))))).))........................................................... (-18.97 = -18.50 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:41 2006