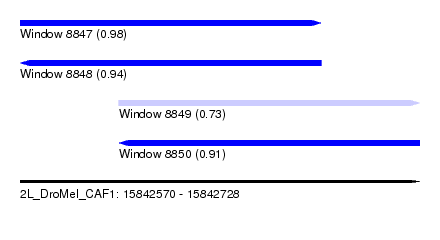
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,842,570 – 15,842,728 |
| Length | 158 |
| Max. P | 0.979913 |

| Location | 15,842,570 – 15,842,689 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.43 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -24.98 |
| Energy contribution | -25.10 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
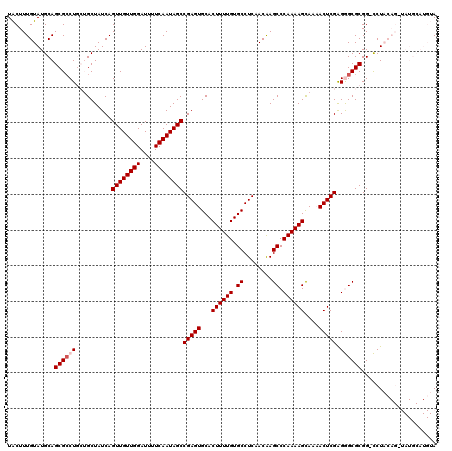
>2L_DroMel_CAF1 15842570 119 + 22407834 UGUGUUAUUCAUCGCUUUUCUAGUUGUUUU-UCCCUCAGCUACUUUGUAUGCAGCGACUGCUGCUAUCAGUUGUUGGAUUUUCAAUAGCCGAGUGCACUUUUGUGCCUCAACAAGCCCAA .............((((.....((((....-(((..(((((.....(((.(((((....)))))))).)))))..)))....))))....(((.((((....)))))))...)))).... ( -31.10) >DroPse_CAF1 1546 114 + 1 U---UUGUCCAUCUAUUUUCUUCUUGUUUUAUCCCUGGGCUGCUUUGUAUGCAGCGCCU---GCUAUCAGUUGUUGGAUUUUCAAUAGCCGAGUGCACUUUUGUGCCUCAACGAGCCCAA .---...............................((((((((((((.((.(((((.((---(....))).))))).))...))).))).(((.((((....)))))))....)))))). ( -31.80) >DroSec_CAF1 14735 119 + 1 UUUGUUAUUCAUCGCUUGUCUAGUUGUUUU-UCCCUCGGCUACUUUGUAUGCAGCGCCUGCUGCUAUCAGUUGUUGGAUUUUCAAUAGCCGAGUGCACUUUUGUGCCUCAACAAGCCCAA .............((((((..((.......-...(((((((...(((...(((((....)))))...)))..(((((....)))))))))))).((((....))))))..)))))).... ( -35.40) >DroSim_CAF1 15036 119 + 1 UGUGUUAUUCAUAGCUUUUCUAGUUGUUUU-UCCCUCGGCUACUUUGUAUGCAGCGCCUGCUGCUAUCAGUUGUUGGAUUUUCAAUAGCCGAGUGCACUUUUGUGCCUCAACAAGCCCAA ..........((((((.....))))))...-......((((....((...(((((....)))))...))((((((((....)))))))).(((.((((....)))))))....))))... ( -32.40) >DroPer_CAF1 27301 114 + 1 U---UUGUCCAUCUAUUUUCUUCUUGUUUUAUCCCUGGGCUGCUUUGUAUGCAGCGCCU---GCUAUCAGUUGUUGGAUUUUCAAUAGCCGAGUGCACUUUUGUGCCUCAACGAGCCCAA .---...............................((((((((((((.((.(((((.((---(....))).))))).))...))).))).(((.((((....)))))))....)))))). ( -31.80) >consensus U_UGUUAUUCAUCGCUUUUCUAGUUGUUUU_UCCCUCGGCUACUUUGUAUGCAGCGCCUGCUGCUAUCAGUUGUUGGAUUUUCAAUAGCCGAGUGCACUUUUGUGCCUCAACAAGCCCAA .......................(((((.........(((((..(((.((.(((((.(((.......))).))))).))...))))))))(((.((((....))))))))))))...... (-24.98 = -25.10 + 0.12)

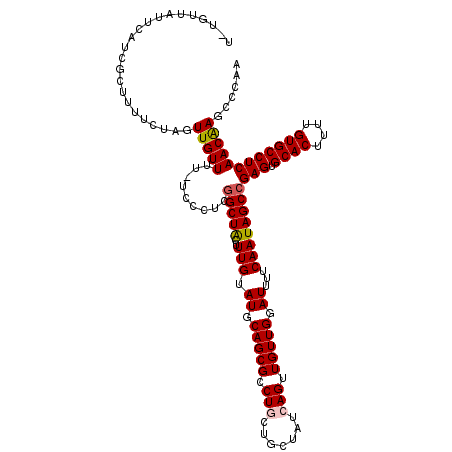
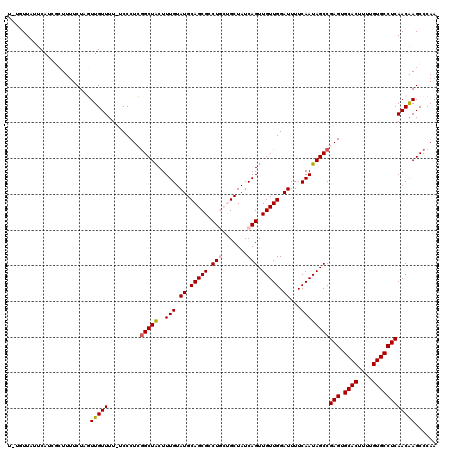
| Location | 15,842,570 – 15,842,689 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.43 |
| Mean single sequence MFE | -34.22 |
| Consensus MFE | -24.06 |
| Energy contribution | -26.30 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
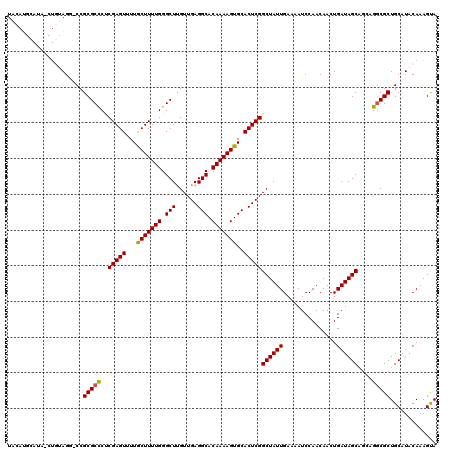
>2L_DroMel_CAF1 15842570 119 - 22407834 UUGGGCUUGUUGAGGCACAAAAGUGCACUCGGCUAUUGAAAAUCCAACAACUGAUAGCAGCAGUCGCUGCAUACAAAGUAGCUGAGGGA-AAAACAACUAGAAAAGCGAUGAAUAACACA ....((((....((((((....)))).((((((((((....(((........))).(((((....)))))......))))))))))...-.......))....))))............. ( -33.70) >DroPse_CAF1 1546 114 - 1 UUGGGCUCGUUGAGGCACAAAAGUGCACUCGGCUAUUGAAAAUCCAACAACUGAUAGC---AGGCGCUGCAUACAAAGCAGCCCAGGGAUAAAACAAGAAGAAAAUAGAUGGACAA---A (((..(((((((((((((....)))).))))))........((((......((....)---)((.(((((.......)))))))..)))).................)).)..)))---. ( -29.60) >DroSec_CAF1 14735 119 - 1 UUGGGCUUGUUGAGGCACAAAAGUGCACUCGGCUAUUGAAAAUCCAACAACUGAUAGCAGCAGGCGCUGCAUACAAAGUAGCCGAGGGA-AAAACAACUAGACAAGCGAUGAAUAACAAA ....(((((((.((((((....)))).((((((((((....(((........))).(((((....)))))......))))))))))...-.......)).)))))))............. ( -41.30) >DroSim_CAF1 15036 119 - 1 UUGGGCUUGUUGAGGCACAAAAGUGCACUCGGCUAUUGAAAAUCCAACAACUGAUAGCAGCAGGCGCUGCAUACAAAGUAGCCGAGGGA-AAAACAACUAGAAAAGCUAUGAAUAACACA ...(((((....((((((....)))).((((((((((....(((........))).(((((....)))))......))))))))))...-.......))....)))))............ ( -36.90) >DroPer_CAF1 27301 114 - 1 UUGGGCUCGUUGAGGCACAAAAGUGCACUCGGCUAUUGAAAAUCCAACAACUGAUAGC---AGGCGCUGCAUACAAAGCAGCCCAGGGAUAAAACAAGAAGAAAAUAGAUGGACAA---A (((..(((((((((((((....)))).))))))........((((......((....)---)((.(((((.......)))))))..)))).................)).)..)))---. ( -29.60) >consensus UUGGGCUUGUUGAGGCACAAAAGUGCACUCGGCUAUUGAAAAUCCAACAACUGAUAGCAGCAGGCGCUGCAUACAAAGUAGCCGAGGGA_AAAACAACUAGAAAAGCGAUGAAUAACA_A .....((.((((..((((....)))).((((((((((....(((........))).(((((....)))))......))))))))))........)))).))................... (-24.06 = -26.30 + 2.24)

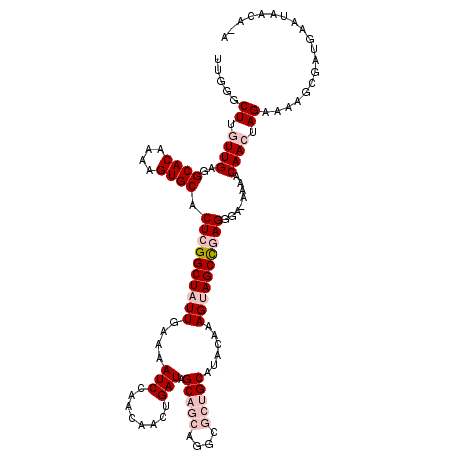

| Location | 15,842,609 – 15,842,728 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.80 |
| Mean single sequence MFE | -37.38 |
| Consensus MFE | -26.80 |
| Energy contribution | -27.40 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15842609 119 + 22407834 UACUUUGUAUGCAGCGACUGCUGCUAUCAGUUGUUGGAUUUUCAAUAGCCGAGUGCACUUUUGUGCCUCAACAAGCCCAAAAGCAAAACUCGGGGGCGCGG-CCUACAGAUAUGCAUGUA (((..(((((((((((((((.......)))))))))...........((((((.((((....))))))).....((((...((.....))...))))..))-).......)))))).))) ( -37.90) >DroPse_CAF1 1583 105 + 1 UGCUUUGUAUGCAGCGCCU---GCUAUCAGUUGUUGGAUUUUCAAUAGCCGAGUGCACUUUUGUGCCUCAACGAGCCCAAAAGUAACACUCGAGAGCGCGGGUCUCG------------A (((.......)))..((((---((..((.((((((((....))))))))((((((.(((((((.((........)).)))))))..)))))).))..))))))....------------. ( -35.00) >DroSec_CAF1 14774 119 + 1 UACUUUGUAUGCAGCGCCUGCUGCUAUCAGUUGUUGGAUUUUCAAUAGCCGAGUGCACUUUUGUGCCUCAACAAGCCCAAAAGCAAAACUCGAGGGCGCGG-CCUACAGCUAUGCAUGUA .......((((((((((((..........((((((((....))))))))(((((...((((((.((........)).))))))....))))).))))))((-(.....))).)))))).. ( -38.90) >DroSim_CAF1 15075 119 + 1 UACUUUGUAUGCAGCGCCUGCUGCUAUCAGUUGUUGGAUUUUCAAUAGCCGAGUGCACUUUUGUGCCUCAACAAGCCCAAAAGCAAAACUCGGGGGCGCGG-CCUACAGAUAUGCAUGUA ...((((((.((.((((((((((....))))((((((....)))))).((((((...((((((.((........)).))))))....)))))))))))).)-).)))))).......... ( -40.10) >DroPer_CAF1 27338 105 + 1 UGCUUUGUAUGCAGCGCCU---GCUAUCAGUUGUUGGAUUUUCAAUAGCCGAGUGCACUUUUGUGCCUCAACGAGCCCAAAAGUAACACUCGAGAGCGCGGGUCUCG------------A (((.......)))..((((---((..((.((((((((....))))))))((((((.(((((((.((........)).)))))))..)))))).))..))))))....------------. ( -35.00) >consensus UACUUUGUAUGCAGCGCCUGCUGCUAUCAGUUGUUGGAUUUUCAAUAGCCGAGUGCACUUUUGUGCCUCAACAAGCCCAAAAGCAAAACUCGAGGGCGCGG_CCUACAG_UAUGCAUGUA .............((((((..........((((((((....))))))))(((((...((((((.((........)).))))))....))))).))))))..................... (-26.80 = -27.40 + 0.60)


| Location | 15,842,609 – 15,842,728 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.80 |
| Mean single sequence MFE | -36.99 |
| Consensus MFE | -28.70 |
| Energy contribution | -28.42 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
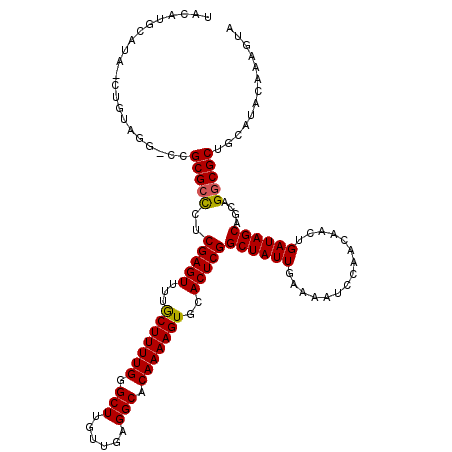
>2L_DroMel_CAF1 15842609 119 - 22407834 UACAUGCAUAUCUGUAGG-CCGCGCCCCCGAGUUUUGCUUUUGGGCUUGUUGAGGCACAAAAGUGCACUCGGCUAUUGAAAAUCCAACAACUGAUAGCAGCAGUCGCUGCAUACAAAGUA ....(((.((((.((.((-((..((((..(((.....)))..)))).....(((((((....)))).))))))).(((......)))..)).))))(((((....))))).......))) ( -37.30) >DroPse_CAF1 1583 105 - 1 U------------CGAGACCCGCGCUCUCGAGUGUUACUUUUGGGCUCGUUGAGGCACAAAAGUGCACUCGGCUAUUGAAAAUCCAACAACUGAUAGC---AGGCGCUGCAUACAAAGCA .------------........(((((..(((((((.((((((((.(((...))).).))))))))))))))((((((...............))))))---.))))).((.......)). ( -35.26) >DroSec_CAF1 14774 119 - 1 UACAUGCAUAGCUGUAGG-CCGCGCCCUCGAGUUUUGCUUUUGGGCUUGUUGAGGCACAAAAGUGCACUCGGCUAUUGAAAAUCCAACAACUGAUAGCAGCAGGCGCUGCAUACAAAGUA ............((((.(-(.(((((..(((((...(((((((.((((....)))).)))))))..)))))((((((...............))))))....))))).)).))))..... ( -38.46) >DroSim_CAF1 15075 119 - 1 UACAUGCAUAUCUGUAGG-CCGCGCCCCCGAGUUUUGCUUUUGGGCUUGUUGAGGCACAAAAGUGCACUCGGCUAUUGAAAAUCCAACAACUGAUAGCAGCAGGCGCUGCAUACAAAGUA ............((((.(-(.(((((..(((((...(((((((.((((....)))).)))))))..)))))((((((...............))))))....))))).)).))))..... ( -38.66) >DroPer_CAF1 27338 105 - 1 U------------CGAGACCCGCGCUCUCGAGUGUUACUUUUGGGCUCGUUGAGGCACAAAAGUGCACUCGGCUAUUGAAAAUCCAACAACUGAUAGC---AGGCGCUGCAUACAAAGCA .------------........(((((..(((((((.((((((((.(((...))).).))))))))))))))((((((...............))))))---.))))).((.......)). ( -35.26) >consensus UACAUGCAUA_CUGUAGG_CCGCGCCCUCGAGUUUUGCUUUUGGGCUUGUUGAGGCACAAAAGUGCACUCGGCUAUUGAAAAUCCAACAACUGAUAGCAGCAGGCGCUGCAUACAAAGUA .....................(((((..(((((...(((((((.(((......))).)))))))..)))))((((((...............))))))....)))))............. (-28.70 = -28.42 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:37 2006