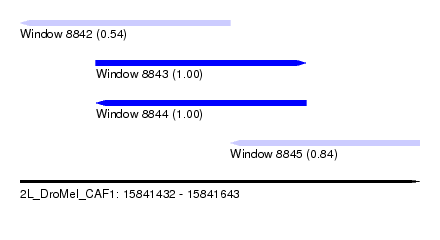
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,841,432 – 15,841,643 |
| Length | 211 |
| Max. P | 0.998510 |

| Location | 15,841,432 – 15,841,543 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.35 |
| Mean single sequence MFE | -14.94 |
| Consensus MFE | -9.07 |
| Energy contribution | -10.07 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15841432 111 - 22407834 CAGUA---AAAAAAAAACGACAAUAAU--ACAUAGCACAAAAAAUGAG----AAAAUUGUCUACAAAAUGGUUGACAGGCAAUUUGCCUACACACAUUUUUCCACCCAAAUGUUUCGUCA .....---..........(((...(((--(.........(((((((..----......(((.((......)).)))((((.....)))).....))))))).........))))..))). ( -15.77) >DroSec_CAF1 13613 109 - 1 CAGUA---AAA--AAAACGACAAUAAU--ACAUAGCACAAAAAAUGAG----AAAAUUGUCUACAAAAUGGUUGACAGGCAAUUUGCCUACACACAUUUUUCCACCCAAAUGUUUCGUCA .....---...--.....(((...(((--(.........(((((((..----......(((.((......)).)))((((.....)))).....))))))).........))))..))). ( -15.77) >DroSim_CAF1 13924 109 - 1 CAGUA---AAA--AAAACGACAAUAAU--ACAUAGCACAAAAAAUGAG----AAAAUUGUCUACAAAAUGGUUGACAGGCAAUUUGCCUACACACAUUUUUCCACCCAAAUGUUUCGUCA .....---...--.....(((...(((--(.........(((((((..----......(((.((......)).)))((((.....)))).....))))))).........))))..))). ( -15.77) >DroPer_CAF1 25992 102 - 1 AAAUACACAAA--AAAGCAAUAAUAACAGACAAAGCAAAAAAAAAAAAAAAAGGAAUUGUCUACAAA-UUUUUGACAGGCAAUUUGCCUACACACAUAUUUGCCC--------------- ...........--...((((.......((((((..(.................)..)))))).....-........((((.....))))..........))))..--------------- ( -12.43) >consensus CAGUA___AAA__AAAACGACAAUAAU__ACAUAGCACAAAAAAUGAG____AAAAUUGUCUACAAAAUGGUUGACAGGCAAUUUGCCUACACACAUUUUUCCACCCAAAUGUUUCGUCA .......................................(((((((............(((.((......)).)))((((.....)))).....)))))))................... ( -9.07 = -10.07 + 1.00)


| Location | 15,841,472 – 15,841,583 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -24.56 |
| Consensus MFE | -17.76 |
| Energy contribution | -17.52 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15841472 111 + 22407834 GCCUGUCAACCAUUUUGUAGACAAUUUU----CUCAUUUUUUGUGCUAUGU--AUUAUUGUCGUUUUUUUUU---UACUGAUGUACAAUGCCUGAUGCUUAUCAAAAAUGGUUGACACAA ...(((((((((((((((((((((....----........)))).))))((--((((((((((((.......---....)))).)))))....)))))......)))))))))))))... ( -26.20) >DroSec_CAF1 13653 109 + 1 GCCUGUCAACCAUUUUGUAGACAAUUUU----CUCAUUUUUUGUGCUAUGU--AUUAUUGUCGUUUU--UUU---UACUGACGUACAUUGCCUGAUGCUUAUCAAAAAUGGUUGACACAA ...(((((((((((((((((((((....----........)))).))))((--((((.(((((((..--...---....)))).))).....))))))......)))))))))))))... ( -27.10) >DroSim_CAF1 13964 109 + 1 GCCUGUCAACCAUUUUGUAGACAAUUUU----CUCAUUUUUUGUGCUAUGU--AUUAUUGUCGUUUU--UUU---UACUGACGUACAUUGCCUGAUGCUUAUCAAAAAUGGUUGACACAA ...(((((((((((((((((((((....----........)))).))))((--((((.(((((((..--...---....)))).))).....))))))......)))))))))))))... ( -27.10) >DroPer_CAF1 26017 111 + 1 GCCUGUCAAAAA-UUUGUAGACAAUUCCUUUUUUUUUUUUUUUUGCUUUGUCUGUUAUUAUUGCUUU--UUUGUGUAUUUCUGU------UCUGAUGUUUAUCAGAAAUGUUUGACGCAA ....(.(((.((-(...(((((((.......................)))))))..))).))))...--.((((((.......(------((((((....))))))).......)))))) ( -17.84) >consensus GCCUGUCAACCAUUUUGUAGACAAUUUU____CUCAUUUUUUGUGCUAUGU__AUUAUUGUCGUUUU__UUU___UACUGACGUACAUUGCCUGAUGCUUAUCAAAAAUGGUUGACACAA ...(((((((((((((...((((((..(......(((.....)))........)..))))))..............................((((....)))))))))))))))))... (-17.76 = -17.52 + -0.25)

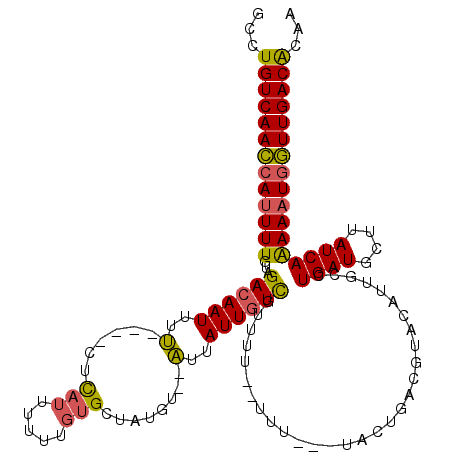
| Location | 15,841,472 – 15,841,583 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -23.56 |
| Consensus MFE | -16.35 |
| Energy contribution | -16.36 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.69 |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.998510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15841472 111 - 22407834 UUGUGUCAACCAUUUUUGAUAAGCAUCAGGCAUUGUACAUCAGUA---AAAAAAAAACGACAAUAAU--ACAUAGCACAAAAAAUGAG----AAAAUUGUCUACAAAAUGGUUGACAGGC .(.(((((((((((((((((..(((........)))..))))...---..........((((((...--.(((..........)))..----...))))))...))))))))))))).). ( -24.20) >DroSec_CAF1 13653 109 - 1 UUGUGUCAACCAUUUUUGAUAAGCAUCAGGCAAUGUACGUCAGUA---AAA--AAAACGACAAUAAU--ACAUAGCACAAAAAAUGAG----AAAAUUGUCUACAAAAUGGUUGACAGGC .(.(((((((((((((.(((((...(((.((.(((((.(((.((.---...--...))))).....)--)))).))........))).----....)))))...))))))))))))).). ( -27.10) >DroSim_CAF1 13964 109 - 1 UUGUGUCAACCAUUUUUGAUAAGCAUCAGGCAAUGUACGUCAGUA---AAA--AAAACGACAAUAAU--ACAUAGCACAAAAAAUGAG----AAAAUUGUCUACAAAAUGGUUGACAGGC .(.(((((((((((((.(((((...(((.((.(((((.(((.((.---...--...))))).....)--)))).))........))).----....)))))...))))))))))))).). ( -27.10) >DroPer_CAF1 26017 111 - 1 UUGCGUCAAACAUUUCUGAUAAACAUCAGA------ACAGAAAUACACAAA--AAAGCAAUAAUAACAGACAAAGCAAAAAAAAAAAAAAAAGGAAUUGUCUACAAA-UUUUUGACAGGC ....((((((...(((((((....))))))------)..............--..............((((((..(.................)..)))))).....-..)))))).... ( -15.83) >consensus UUGUGUCAACCAUUUUUGAUAAGCAUCAGGCAAUGUACAUCAGUA___AAA__AAAACGACAAUAAU__ACAUAGCACAAAAAAUGAG____AAAAUUGUCUACAAAAUGGUUGACAGGC ....((((((((((((((((....))))).............................((((((...............................))))))....))))))))))).... (-16.35 = -16.36 + 0.00)


| Location | 15,841,543 – 15,841,643 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -18.60 |
| Consensus MFE | -17.91 |
| Energy contribution | -17.80 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15841543 100 - 22407834 ACUGUCGCAUUGUACACAACCUAUUGCAAAUAAACACAUCAUACGUUUUUAUGACGAAAAUUGUGUCAACCAUUUUUGAUAAGCAUCAGGCAUUGUACAU ..........((((((...(((..(((...........(((((......)))))(((((((.((....)).)))))))....)))..)))...)))))). ( -21.00) >DroSec_CAF1 13722 100 - 1 ACUAUCGCAUUGUACACAACUUAUUGCAAAUAAACACAUCAUACGUUUUUAUUACGAAAAUUGUGUCAACCAUUUUUGAUAAGCAUCAGGCAAUGUACGU ......(((((((......(((((((.((((..(((((.....(((.......))).....))))).....)))).)))))))......))))))).... ( -17.50) >DroSim_CAF1 14033 100 - 1 ACUAUCGAAUUGUACACAACUUAUUGCAAAUAAACACAUCAUACGUUUUUAUUACGAAAAUUGUGUCAACCAUUUUUGAUAAGCAUCAGGCAAUGUACGU ...........(((((...(((..(((.((((((.((.......)).)))))).(((((((.((....)).)))))))....)))..)))...))))).. ( -17.30) >consensus ACUAUCGCAUUGUACACAACUUAUUGCAAAUAAACACAUCAUACGUUUUUAUUACGAAAAUUGUGUCAACCAUUUUUGAUAAGCAUCAGGCAAUGUACGU ..........((((((...(((..(((.((((((.((.......)).)))))).(((((((.((....)).)))))))....)))..)))...)))))). (-17.91 = -17.80 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:32 2006