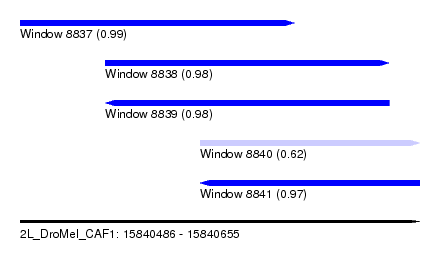
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,840,486 – 15,840,655 |
| Length | 169 |
| Max. P | 0.988460 |

| Location | 15,840,486 – 15,840,602 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.71 |
| Mean single sequence MFE | -37.36 |
| Consensus MFE | -31.50 |
| Energy contribution | -33.38 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15840486 116 + 22407834 ---GUGCUGUGGCUAGA-AGCCAGCGAAGCAAAAUGCUUUUAAUUAAAAGUAAACAAUAUCAAUUCGUUCGACAGUUAUCCGGUGAUGCUGCCAGCAUUGCCGGGUGUCGAGCAUCCAGG ---.((((.((((....-.))))....))))...(((((((....)))))))..............((((((((....(((((..((((.....))))..)))))))))))))....... ( -42.30) >DroSec_CAF1 12522 116 + 1 ---GUGCUGUGGCUAGA-AGCCAGCGAAGCAAAUUGCUUUUAAUUAAAAGUAAACAAUAUCAAUUCGUUCGACAGUUAUCCGGUGAUGCUGCCAGCAUUGCCGGGUGUCGAGCAUCCAGA ---.((((.((((....-.))))....))))..((((((((....)))))))).............((((((((....(((((..((((.....))))..)))))))))))))....... ( -43.20) >DroSim_CAF1 12832 116 + 1 ---GUGCUGUGGCUAGA-AGCCAGCGAAGCAAAAUGCUUUUAAUUAAAAGUAAACAAUAUCAAUUCGUACGACAGUUAUCCGGUGAUGCUGCCAGCAUUGCCGGGUGUCGAGCAUCCAGG ---(((((.((((....-.))))(((((......(((((((....)))))))...........))))).(((((....(((((..((((.....))))..)))))))))))))))..... ( -38.83) >DroPer_CAF1 21549 111 + 1 UCAGUGCUGUGGCAAGAAAGCCAGCGAAGCAAAUUGUUUUUAAUUAAAAGUAAACAAUAUCAAUUCAUUCGACAGUUAUCCUGUUAUGCUGCUGACAUUACUGGCUGGCG---------C ....(((....))).....(((((((((....(((((((............))))))).....)))......((((.....(((((......)))))..)))))))))).---------. ( -25.10) >consensus ___GUGCUGUGGCUAGA_AGCCAGCGAAGCAAAAUGCUUUUAAUUAAAAGUAAACAAUAUCAAUUCGUUCGACAGUUAUCCGGUGAUGCUGCCAGCAUUGCCGGGUGUCGAGCAUCCAGG ....((((.((((......))))....))))...(((((((....)))))))..............((((((((....(((((((((((.....)))))))))))))))))))....... (-31.50 = -33.38 + 1.88)

| Location | 15,840,522 – 15,840,642 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.00 |
| Mean single sequence MFE | -42.17 |
| Consensus MFE | -29.55 |
| Energy contribution | -33.43 |
| Covariance contribution | 3.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15840522 120 + 22407834 UAAUUAAAAGUAAACAAUAUCAAUUCGUUCGACAGUUAUCCGGUGAUGCUGCCAGCAUUGCCGGGUGUCGAGCAUCCAGGCGUGGAGCCUGACGUGGCCAGGACCAAGCCAGGAUGCAUU ............................((((((....(((((..((((.....))))..)))))))))))((((((.(((.(((..((((.......)))).))).))).))))))... ( -50.10) >DroSec_CAF1 12558 120 + 1 UAAUUAAAAGUAAACAAUAUCAAUUCGUUCGACAGUUAUCCGGUGAUGCUGCCAGCAUUGCCGGGUGUCGAGCAUCCAGACGUGGAGCCUGACGUGGCCAGGACCAAGUCAGGAUGCAUU ............................((((((....(((((..((((.....))))..)))))))))))((((((.(((.(((..((((.......)))).))).))).))))))... ( -48.00) >DroSim_CAF1 12868 120 + 1 UAAUUAAAAGUAAACAAUAUCAAUUCGUACGACAGUUAUCCGGUGAUGCUGCCAGCAUUGCCGGGUGUCGAGCAUCCAGGCGUGGAGCCUGACGUGGCCAGGACCAAGCCAGGAUGCAUU .............................(((((....(((((..((((.....))))..)))))))))).((((((.(((.(((..((((.......)))).))).))).))))))... ( -48.80) >DroPer_CAF1 21589 110 + 1 UAAUUAAAAGUAAACAAUAUCAAUUCAUUCGACAGUUAUCCUGUUAUGCUGCUGACAUUACUGGCUGGCG---------CCAUAGACGGUGACGUGUCCAGGACCUCCUCU-AUUGCAUU .........((((.................((((((((((...(((((.((((..(.......)..))))---------.)))))..)))))).)))).(((....)))..-.))))... ( -21.80) >consensus UAAUUAAAAGUAAACAAUAUCAAUUCGUUCGACAGUUAUCCGGUGAUGCUGCCAGCAUUGCCGGGUGUCGAGCAUCCAGGCGUGGAGCCUGACGUGGCCAGGACCAAGCCAGGAUGCAUU .............................(((((....(((((((((((.....)))))))))))))))).((((((.(((.(((..((((.......)))).))).))).))))))... (-29.55 = -33.43 + 3.88)

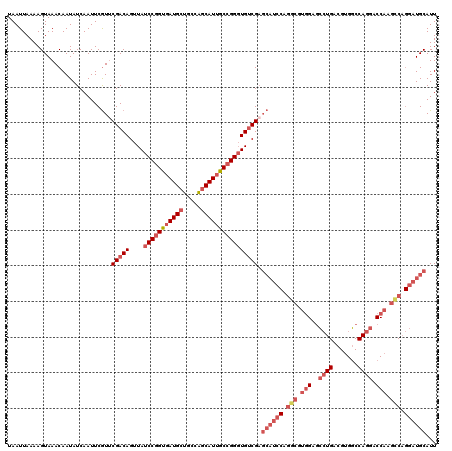
| Location | 15,840,522 – 15,840,642 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.00 |
| Mean single sequence MFE | -40.00 |
| Consensus MFE | -28.41 |
| Energy contribution | -31.35 |
| Covariance contribution | 2.94 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.977023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15840522 120 - 22407834 AAUGCAUCCUGGCUUGGUCCUGGCCACGUCAGGCUCCACGCCUGGAUGCUCGACACCCGGCAAUGCUGGCAGCAUCACCGGAUAACUGUCGAACGAAUUGAUAUUGUUUACUUUUAAUUA ...((((((.(((.(((.((((((...))))))..))).))).))))))((((((.((((..((((.....))))..)))).....))))))...((((((............)))))). ( -46.30) >DroSec_CAF1 12558 120 - 1 AAUGCAUCCUGACUUGGUCCUGGCCACGUCAGGCUCCACGUCUGGAUGCUCGACACCCGGCAAUGCUGGCAGCAUCACCGGAUAACUGUCGAACGAAUUGAUAUUGUUUACUUUUAAUUA ...((((((.(((.(((.((((((...))))))..))).))).))))))((((((.((((..((((.....))))..)))).....))))))...((((((............)))))). ( -44.20) >DroSim_CAF1 12868 120 - 1 AAUGCAUCCUGGCUUGGUCCUGGCCACGUCAGGCUCCACGCCUGGAUGCUCGACACCCGGCAAUGCUGGCAGCAUCACCGGAUAACUGUCGUACGAAUUGAUAUUGUUUACUUUUAAUUA ...((((((.(((.(((.((((((...))))))..))).))).)))))).(((((.((((..((((.....))))..)))).....)))))....((((((............)))))). ( -44.20) >DroPer_CAF1 21589 110 - 1 AAUGCAAU-AGAGGAGGUCCUGGACACGUCACCGUCUAUGG---------CGCCAGCCAGUAAUGUCAGCAGCAUAACAGGAUAACUGUCGAAUGAAUUGAUAUUGUUUACUUUUAAUUA .......(-(((((.((((((((((........))))).))---------.)))......(((((((((...(((.((((.....))))...)))..)))))))))....)))))).... ( -25.30) >consensus AAUGCAUCCUGACUUGGUCCUGGCCACGUCAGGCUCCACGCCUGGAUGCUCGACACCCGGCAAUGCUGGCAGCAUCACCGGAUAACUGUCGAACGAAUUGAUAUUGUUUACUUUUAAUUA ...((((((.(((.(((.((((((...))))))..))).))).)))))).(((((.((((..((((.....))))..)))).....)))))....((((((............)))))). (-28.41 = -31.35 + 2.94)

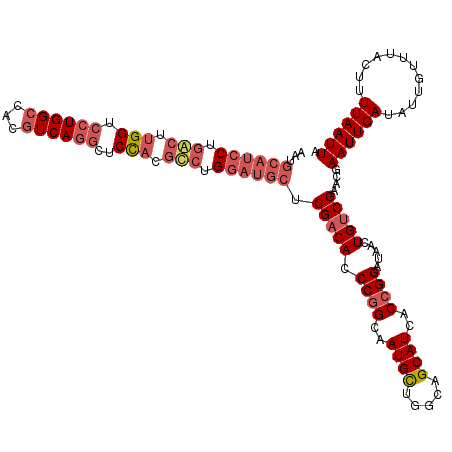
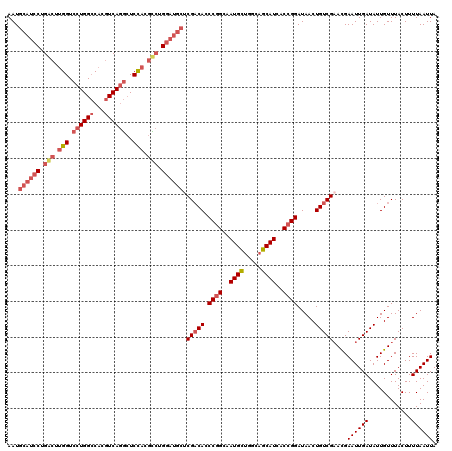
| Location | 15,840,562 – 15,840,655 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 98.57 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -41.04 |
| Energy contribution | -40.60 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.42 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15840562 93 + 22407834 CGGUGAUGCUGCCAGCAUUGCCGGGUGUCGAGCAUCCAGGCGUGGAGCCUGACGUGGCCAGGACCAAGCCAGGAUGCAUUGUUCUCGGGGGAG .((..((((.....))))..))(((...(((((((((.(((.(((..((((.......)))).))).))).)))))).)))..)))....... ( -41.30) >DroSec_CAF1 12598 93 + 1 CGGUGAUGCUGCCAGCAUUGCCGGGUGUCGAGCAUCCAGACGUGGAGCCUGACGUGGCCAGGACCAAGUCAGGAUGCAUUGUUCUCGGGGGAG .((..((((.....))))..))(((...(((((((((.(((.(((..((((.......)))).))).))).)))))).)))..)))....... ( -39.20) >DroSim_CAF1 12908 93 + 1 CGGUGAUGCUGCCAGCAUUGCCGGGUGUCGAGCAUCCAGGCGUGGAGCCUGACGUGGCCAGGACCAAGCCAGGAUGCAUUGUUCUCGGGGGAG .((..((((.....))))..))(((...(((((((((.(((.(((..((((.......)))).))).))).)))))).)))..)))....... ( -41.30) >consensus CGGUGAUGCUGCCAGCAUUGCCGGGUGUCGAGCAUCCAGGCGUGGAGCCUGACGUGGCCAGGACCAAGCCAGGAUGCAUUGUUCUCGGGGGAG .((..((((.....))))..))(((...(((((((((.(((.(((..((((.......)))).))).))).)))))).)))..)))....... (-41.04 = -40.60 + -0.44)

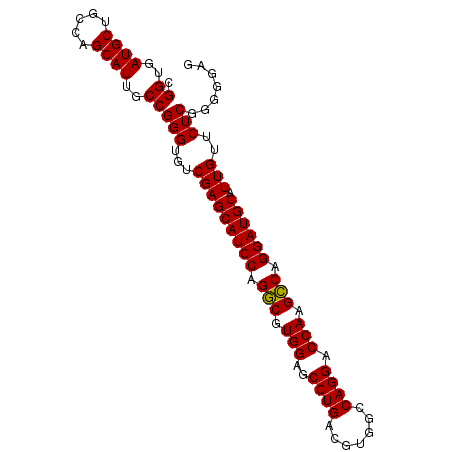
| Location | 15,840,562 – 15,840,655 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 98.57 |
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -36.54 |
| Energy contribution | -36.10 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.35 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15840562 93 - 22407834 CUCCCCCGAGAACAAUGCAUCCUGGCUUGGUCCUGGCCACGUCAGGCUCCACGCCUGGAUGCUCGACACCCGGCAAUGCUGGCAGCAUCACCG (((....)))......((((((.(((.(((.((((((...))))))..))).))).))))))........(((..((((.....))))..))) ( -36.80) >DroSec_CAF1 12598 93 - 1 CUCCCCCGAGAACAAUGCAUCCUGACUUGGUCCUGGCCACGUCAGGCUCCACGUCUGGAUGCUCGACACCCGGCAAUGCUGGCAGCAUCACCG (((....)))......((((((.(((.(((.((((((...))))))..))).))).))))))........(((..((((.....))))..))) ( -34.70) >DroSim_CAF1 12908 93 - 1 CUCCCCCGAGAACAAUGCAUCCUGGCUUGGUCCUGGCCACGUCAGGCUCCACGCCUGGAUGCUCGACACCCGGCAAUGCUGGCAGCAUCACCG (((....)))......((((((.(((.(((.((((((...))))))..))).))).))))))........(((..((((.....))))..))) ( -36.80) >consensus CUCCCCCGAGAACAAUGCAUCCUGGCUUGGUCCUGGCCACGUCAGGCUCCACGCCUGGAUGCUCGACACCCGGCAAUGCUGGCAGCAUCACCG (((....)))......((((((.(((.(((.((((((...))))))..))).))).))))))........(((..((((.....))))..))) (-36.54 = -36.10 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:28 2006