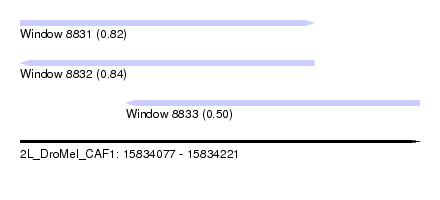
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,834,077 – 15,834,221 |
| Length | 144 |
| Max. P | 0.841741 |

| Location | 15,834,077 – 15,834,183 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.55 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -18.02 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15834077 106 + 22407834 ACAAGCAUCUGGAUACACACAUUUAUAU--AGUACACAUCUGUGUGUAUAUGUA-----------UAUGGAGUGGCACAACACAUGCUUCGACUGC-UUCGCAGCUACGCUUUGCUGCCU ..((((((.((....(((.(((.(((((--(.((((((....))))))))))))-----------.)))..)))......)).)))))).......-...(((((........))))).. ( -30.60) >DroSec_CAF1 7212 108 + 1 ACAAGCAUCUGGAUACACACUUUUAUAU--AGUACACAUCUGUAUGUAUAUGUA---------UAUAUGGAGUGGCACAACACAUGCUUCGACUGC-UUCGCAGCUACGCUUUGCUGCCU ..((((((.((......(((((((((((--((((.((((....)))).))).))---------)))).))))))......)).)))))).......-...(((((........))))).. ( -26.60) >DroSim_CAF1 7401 106 + 1 ACAAGCAUUUGGAUACACACUUUUAUAU--AGUACACAUCUGUGUGUAUAUGUA-----------UAUGGAGUGGCACAACACAUGCUUCGACUGC-UUCGCAGCUACGCUUUGCUGCCU ..(((((.(((((....(((((((((((--((((((((....))))))).))))-----------)).))))))(((.......)))))))).)))-)).(((((........))))).. ( -33.20) >DroPer_CAF1 15279 118 + 1 ACAUCUAUCUAGAUACAAACGCACAGAACAAGUA--CAUAUGUAUAUAUAAGUAUGGUGUCUCUAUAUGGAGUGGCACAACAUAUGCUUGAGCUGCCGUCGCAGCCACGCUUUGCUGCCU ..((((....))))....(((..(((..((((..--(((((((((((....)))).(((((.((......)).))))).)))))))))))..))).))).(((((........))))).. ( -29.50) >consensus ACAAGCAUCUGGAUACACACUUUUAUAU__AGUACACAUCUGUAUGUAUAUGUA___________UAUGGAGUGGCACAACACAUGCUUCGACUGC_UUCGCAGCUACGCUUUGCUGCCU ..((((((....((((((((.....................))))))))......................(((......)))))))))...........(((((........))))).. (-18.02 = -18.90 + 0.88)

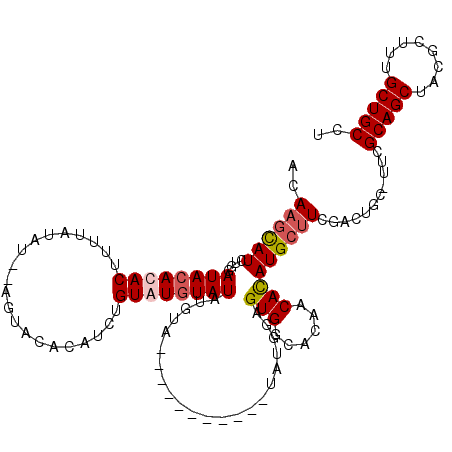

| Location | 15,834,077 – 15,834,183 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.55 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -22.29 |
| Energy contribution | -22.60 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15834077 106 - 22407834 AGGCAGCAAAGCGUAGCUGCGAA-GCAGUCGAAGCAUGUGUUGUGCCACUCCAUA-----------UACAUAUACACACAGAUGUGUACU--AUAUAAAUGUGUGUAUCCAGAUGCUUGU ..(((((........))))).((-(((.((.....((((((.(((......))))-----------)))))(((((((((..(((((...--)))))..)))))))))...))))))).. ( -31.20) >DroSec_CAF1 7212 108 - 1 AGGCAGCAAAGCGUAGCUGCGAA-GCAGUCGAAGCAUGUGUUGUGCCACUCCAUAUA---------UACAUAUACAUACAGAUGUGUACU--AUAUAAAAGUGUGUAUCCAGAUGCUUGU ..(((((........)))))...-.......((((((.((..((((((((...((((---------((..(((((((....))))))).)--)))))..)))).)))).)).)))))).. ( -31.90) >DroSim_CAF1 7401 106 - 1 AGGCAGCAAAGCGUAGCUGCGAA-GCAGUCGAAGCAUGUGUUGUGCCACUCCAUA-----------UACAUAUACACACAGAUGUGUACU--AUAUAAAAGUGUGUAUCCAAAUGCUUGU ..(((((........)))))...-.......((((((.((..((((((((..(((-----------((....((((((....)))))).)--))))...)))).)))).)).)))))).. ( -27.80) >DroPer_CAF1 15279 118 - 1 AGGCAGCAAAGCGUGGCUGCGACGGCAGCUCAAGCAUAUGUUGUGCCACUCCAUAUAGAGACACCAUACUUAUAUAUACAUAUG--UACUUGUUCUGUGCGUUUGUAUCUAGAUAGAUGU .(((((((..((.(((((((....))))).)).))...)))..)))).(((......)))....(((.((..((.(((((.(((--(((.......)))))).))))).))...))))). ( -33.40) >consensus AGGCAGCAAAGCGUAGCUGCGAA_GCAGUCGAAGCAUGUGUUGUGCCACUCCAUA___________UACAUAUACACACAGAUGUGUACU__AUAUAAAAGUGUGUAUCCAGAUGCUUGU .(((((((..((...(((((....)))))....))...)))..))))........................((((((((...(((((.....)))))...))))))))............ (-22.29 = -22.60 + 0.31)



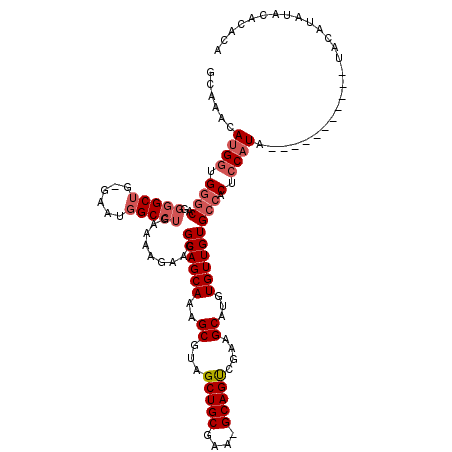
| Location | 15,834,115 – 15,834,221 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.09 |
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -24.16 |
| Energy contribution | -25.73 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15834115 106 - 22407834 GCAAACAUGGAGGGCAGGGGCUG--AAUGGCCUGAAAAGAAGGCAGCAAAGCGUAGCUGCGAA-GCAGUCGAAGCAUGUGUUGUGCCACUCCAUA-----------UACAUAUACACACA ......(((((((((..(((((.--...))))).........((((((..((...(((((...-)))))....))...))))))))).)))))).-----------.............. ( -35.40) >DroSec_CAF1 7250 109 - 1 GCAAACAUGGUGGGCAGGGGCUG-GAAUGGCCUGAAAAGAAGGCAGCAAAGCGUAGCUGCGAA-GCAGUCGAAGCAUGUGUUGUGCCACUCCAUAUA---------UACAUAUACAUACA ......((((.((((..(((((.-....))))).........((((((..((...(((((...-)))))....))...))))))))).).))))...---------.............. ( -31.20) >DroSim_CAF1 7439 107 - 1 GCAAACAUGGUGGGCAGGGGCUG-GAAUGGCCUGAAAAGAAGGCAGCAAAGCGUAGCUGCGAA-GCAGUCGAAGCAUGUGUUGUGCCACUCCAUA-----------UACAUAUACACACA ......((((.((((..(((((.-....))))).........((((((..((...(((((...-)))))....))...))))))))).).)))).-----------.............. ( -31.20) >DroPer_CAF1 15317 119 - 1 GCAAACAAG-CGCUCGGAGGCGGCCAAGGGCGAGAAAAGAAGGCAGCAAAGCGUGGCUGCGACGGCAGCUCAAGCAUAUGUUGUGCCACUCCAUAUAGAGACACCAUACUUAUAUAUACA ((......)-)....((.(((.(((...)))...........((((((..((.(((((((....))))).)).))...))))))))).(((......)))...))............... ( -31.90) >consensus GCAAACAUGGUGGGCAGGGGCUG_GAAUGGCCUGAAAAGAAGGCAGCAAAGCGUAGCUGCGAA_GCAGUCGAAGCAUGUGUUGUGCCACUCCAUA___________UACAUAUACACACA ......((((.((((..(((((......))))).........((((((..((...(((((....)))))....))...))))))))).).)))).......................... (-24.16 = -25.73 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:21 2006