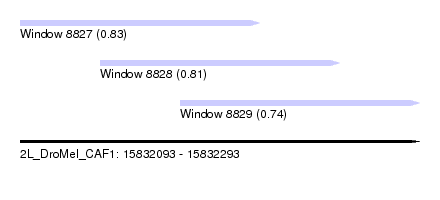
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,832,093 – 15,832,293 |
| Length | 200 |
| Max. P | 0.831290 |

| Location | 15,832,093 – 15,832,213 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -36.83 |
| Consensus MFE | -33.52 |
| Energy contribution | -34.30 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15832093 120 + 22407834 AAACUUUGCCAGCUGUAUUGCAUUACGCCUUUAAUUUCAUUACACGUCCAGACGGAACGGGAAAACUCCCGCCCAAGUUCCGAGCUGGCCCAUAAAGUCGAGGGAAACACUUGUAAAGUC ..(((((((((((((((.((.((((......))))..)).))).........((((((((((....))))......))))))))))))).........(((((....).)))).))))). ( -33.30) >DroSec_CAF1 4687 120 + 1 AAACUUUGCCAGCUGUAUUGCAUUACGCCCUUAAUUUCAUUACACGGCCAGUGGGAACGGGGAAAGUCCCGCCCAAGUUCCGAGCUGGCCCAUAAAAUCGAGGGAAACACUUGUAAAGUC ..(((((((.((..(((......))).((((..((((.((.....(((((((.(((((.(((.........)))..)))))..))))))).)).))))..)))).....)).))))))). ( -39.50) >DroSim_CAF1 4876 120 + 1 AAACUUUGCCAGCUGUAUUGCAUUACGCCUUUAAUUUCAUUACACGGCCAGUGGGAACGGGGAAACUCCCGCCCAAGUUCCGAGCUGGCCCAUAAAAUCGAGGGAAACACUUGUAAAGUC ..(((((((((((((((.((.((((......))))..)).))).(((....((((..(((((....)))))))))....)))))))))).........(((((....).)))).))))). ( -37.70) >consensus AAACUUUGCCAGCUGUAUUGCAUUACGCCUUUAAUUUCAUUACACGGCCAGUGGGAACGGGGAAACUCCCGCCCAAGUUCCGAGCUGGCCCAUAAAAUCGAGGGAAACACUUGUAAAGUC ..(((((((((((((((.((.((((......))))..)).))).(((....((((..(((((....)))))))))....)))))))))).........(((((....).)))).))))). (-33.52 = -34.30 + 0.78)

| Location | 15,832,133 – 15,832,253 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -44.81 |
| Consensus MFE | -39.59 |
| Energy contribution | -40.37 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15832133 120 + 22407834 UACACGUCCAGACGGAACGGGAAAACUCCCGCCCAAGUUCCGAGCUGGCCCAUAAAGUCGAGGGAAACACUUGUAAAGUCUGCCGGGCGACCAGAAAGGGAGCCCUUUCCUAUGGUCUCA .....((((...((((((((((....))))......))))))....(((.........(((((....).))))........)))))))(((((((((((....))))))...)))))... ( -43.33) >DroSec_CAF1 4727 120 + 1 UACACGGCCAGUGGGAACGGGGAAAGUCCCGCCCAAGUUCCGAGCUGGCCCAUAAAAUCGAGGGAAACACUUGUAAAGUCUGCCGGGCGACCAGAAAGGGAGCCCUUUCCUAUGGUCUCA .....(((((((.(((((.(((.........)))..)))))..)))))))........(((((....).))))....(((.....)))(((((((((((....))))))...)))))... ( -46.40) >DroSim_CAF1 4916 120 + 1 UACACGGCCAGUGGGAACGGGGAAACUCCCGCCCAAGUUCCGAGCUGGCCCAUAAAAUCGAGGGAAACACUUGUAAAGUCUGCCGGGCGACCAGAAAGGGAGCCCUUUCCAAAGGUCUCA .....(((((((.(((((.(((.........)))..)))))..)))))))........(((((....).))))....(((.....)))((((.((((((....))))))....))))... ( -44.70) >consensus UACACGGCCAGUGGGAACGGGGAAACUCCCGCCCAAGUUCCGAGCUGGCCCAUAAAAUCGAGGGAAACACUUGUAAAGUCUGCCGGGCGACCAGAAAGGGAGCCCUUUCCUAUGGUCUCA .....(((((((.(((((((((....))))......)))))..)))))))........(((((....).))))....(((.....)))(((((((((((....))))))...)))))... (-39.59 = -40.37 + 0.78)


| Location | 15,832,173 – 15,832,293 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -41.51 |
| Consensus MFE | -38.80 |
| Energy contribution | -39.47 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15832173 120 + 22407834 CGAGCUGGCCCAUAAAGUCGAGGGAAACACUUGUAAAGUCUGCCGGGCGACCAGAAAGGGAGCCCUUUCCUAUGGUCUCAUAUUGCAUGGCGCCUCAUCUCGACCAACACUGCAGCUCAG .((((((.(.......(((((((((..(.(((((((.(((.....)))(((((((((((....))))))...))))).....))))).)).)..)).))))))).......))))))).. ( -41.44) >DroSec_CAF1 4767 120 + 1 CGAGCUGGCCCAUAAAAUCGAGGGAAACACUUGUAAAGUCUGCCGGGCGACCAGAAAGGGAGCCCUUUCCUAUGGUCUCAUAUUGCAUGGCGCCUCACCUCGAGCAACACUGCAGCUCAG .((((((.(........((((((((..(.(((((((.(((.....)))(((((((((((....))))))...))))).....))))).)).)..)).))))))........))))))).. ( -42.49) >DroSim_CAF1 4956 120 + 1 CGAGCUGGCCCAUAAAAUCGAGGGAAACACUUGUAAAGUCUGCCGGGCGACCAGAAAGGGAGCCCUUUCCAAAGGUCUCAUAUUGCAUGGCGCCUCAUCUCCAGCAACACUGCAGCUCAG .((((((((((........((((....).)))(((.....))).))))((((.((((((....))))))....)))).....((((.(((.(......).))))))).....)))))).. ( -40.60) >consensus CGAGCUGGCCCAUAAAAUCGAGGGAAACACUUGUAAAGUCUGCCGGGCGACCAGAAAGGGAGCCCUUUCCUAUGGUCUCAUAUUGCAUGGCGCCUCAUCUCGAGCAACACUGCAGCUCAG .((((((((((((......((((....).)))((((.(((.....)))(((((((((((....))))))...))))).....)))))))).))).........(((....)))))))).. (-38.80 = -39.47 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:17 2006