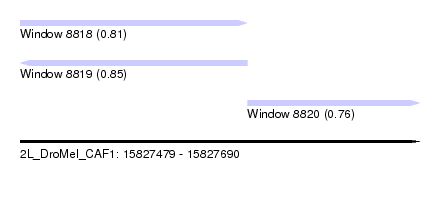
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,827,479 – 15,827,690 |
| Length | 211 |
| Max. P | 0.851519 |

| Location | 15,827,479 – 15,827,599 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -45.50 |
| Consensus MFE | -41.69 |
| Energy contribution | -42.13 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15827479 120 + 22407834 ACAGACUGAGAACGGGUGGGUGGGCGCUAGAUGGCGAGGUGGCAAGGACAUCCUUUUCCAUGGCAGCCGGCAAAUAUCGAGAGGUAAACACGUGCAUUGCCAGCUUCCGUUAUCAGUCAG ...((((((.((((((..((..(((((..(.(((((..((((.((((....))))..))))..).)))).)...((((....)))).....)))).)..))....)))))).)))))).. ( -46.90) >DroSec_CAF1 15 119 + 1 ACAGACUGAGAACGGGUGGGUG-GCGCUAGAAGGCGAGGUGGCGAGGACAUCCUUUUCCAUGGCAGCCGGCAAAUAUCGAGAGGUAAACACGAGCAUUGCCAGCUUCCGUUAUCAGUCAG ...((((((.((((((.((.((-(((((.(..((((..((((.((((....))))..))))..).)))..)...((((....))))......)))...)))).)))))))).)))))).. ( -46.60) >DroSim_CAF1 201 120 + 1 ACAUACUGAGAACGGGUGGGUGGGCGCUAGAAGGCGAGGUGGCGAGGACAUCCUUUUCCAUGGCAGCCGGCAAAUAUCGAGAGGUAAACACGUGCAUUGCCAGCUUCCGUUAUCAGUCAG ....(((((.((((((..((..(((((..(..((((..((((.((((....))))..))))..).)))..)...((((....)))).....)))).)..))....)))))).)))))... ( -43.00) >consensus ACAGACUGAGAACGGGUGGGUGGGCGCUAGAAGGCGAGGUGGCGAGGACAUCCUUUUCCAUGGCAGCCGGCAAAUAUCGAGAGGUAAACACGUGCAUUGCCAGCUUCCGUUAUCAGUCAG ...((((((.((((((..(((..((((..(..((((..((((.((((....))))..))))..).)))..)...((((....)))).....))))...)))....)))))).)))))).. (-41.69 = -42.13 + 0.45)

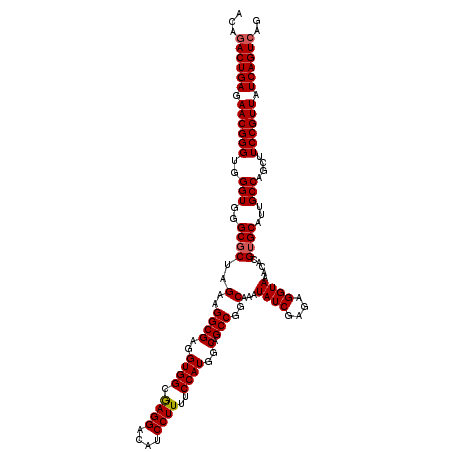
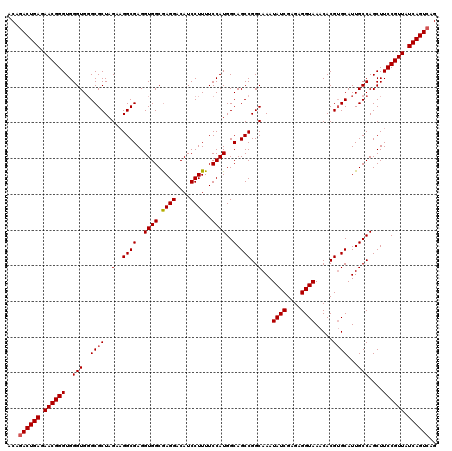
| Location | 15,827,479 – 15,827,599 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -38.67 |
| Consensus MFE | -37.00 |
| Energy contribution | -37.33 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15827479 120 - 22407834 CUGACUGAUAACGGAAGCUGGCAAUGCACGUGUUUACCUCUCGAUAUUUGCCGGCUGCCAUGGAAAAGGAUGUCCUUGCCACCUCGCCAUCUAGCGCCCACCCACCCGUUCUCAGUCUGU ..((((((.(((((.(((((((((....((.(.......).))....)))))))))((..(((..((((....)))).)))....((......))))........))))).))))))... ( -39.20) >DroSec_CAF1 15 119 - 1 CUGACUGAUAACGGAAGCUGGCAAUGCUCGUGUUUACCUCUCGAUAUUUGCCGGCUGCCAUGGAAAAGGAUGUCCUCGCCACCUCGCCUUCUAGCGC-CACCCACCCGUUCUCAGUCUGU ..((((((.(((((.(((((((((...(((.(.......).)))...)))))))))((..(((...(((....)))..)))....((......))))-.......))))).))))))... ( -40.90) >DroSim_CAF1 201 120 - 1 CUGACUGAUAACGGAAGCUGGCAAUGCACGUGUUUACCUCUCGAUAUUUGCCGGCUGCCAUGGAAAAGGAUGUCCUCGCCACCUCGCCUUCUAGCGCCCACCCACCCGUUCUCAGUAUGU ...(((((.(((((.(((((((((....((.(.......).))....)))))))))((..(((...(((....)))..)))....((......))))........))))).))))).... ( -35.90) >consensus CUGACUGAUAACGGAAGCUGGCAAUGCACGUGUUUACCUCUCGAUAUUUGCCGGCUGCCAUGGAAAAGGAUGUCCUCGCCACCUCGCCUUCUAGCGCCCACCCACCCGUUCUCAGUCUGU ..((((((.(((((.(((((((((....((.(.......).))....)))))))))....(((...(((....)))..)))...(((......))).........))))).))))))... (-37.00 = -37.33 + 0.33)

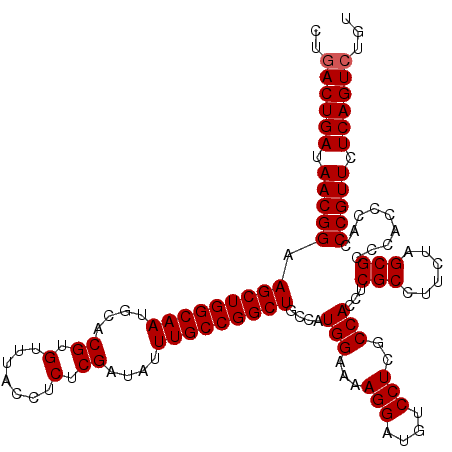

| Location | 15,827,599 – 15,827,690 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 98.53 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -25.62 |
| Energy contribution | -25.40 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.60 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15827599 91 + 22407834 CUUGUUUGCCAGACAGCCGGGCCAUUUCCAUUGGACGCGCCCACCCGGUUUACCUGGCCAACAUCGCUCCCCCAUCGCCGUUCGUCUAUAU ..((((.(((((..(((((((..........(((......))))))))))...))))).))))............................ ( -23.80) >DroSec_CAF1 134 91 + 1 CUUGUUUGCCAGACAGCCGGGCCAUUUCCAUUGGACGCGCCCACCCGGCUUACCUGGCCAACAUCGCUCCCCCAUCGCUGUUCGUCUAUAU ..((((.(((((..(((((((..........(((......))))))))))...))))).))))............................ ( -26.20) >DroSim_CAF1 321 91 + 1 CUUGUUUGCCAGACAGCCGGGCCAUUUCCAUUGGACGCGCCCACCCGGCUUACCUGGCCAACAUCGCUCCCCCAUCGCUGUUCGUCUAUAU ..((((.(((((..(((((((..........(((......))))))))))...))))).))))............................ ( -26.20) >consensus CUUGUUUGCCAGACAGCCGGGCCAUUUCCAUUGGACGCGCCCACCCGGCUUACCUGGCCAACAUCGCUCCCCCAUCGCUGUUCGUCUAUAU ..((((.(((((..(((((((..........(((......))))))))))...))))).))))............................ (-25.62 = -25.40 + -0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:09 2006