
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
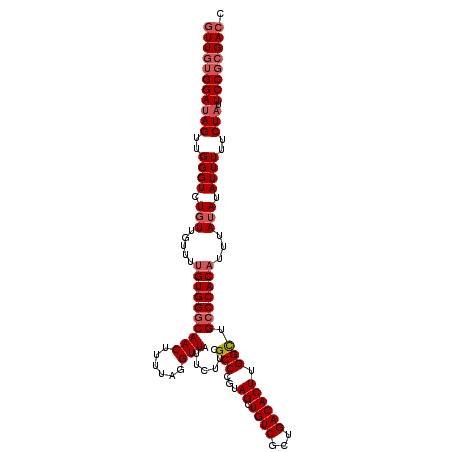
| Location | 15,826,143 – 15,826,255 |
| Length | 112 |
| Max. P | 0.912942 |

| Location | 15,826,143 – 15,826,255 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 94.05 |
| Mean single sequence MFE | -33.51 |
| Consensus MFE | -28.14 |
| Energy contribution | -29.70 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.912942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15826143 112 + 22407834 GUUGUGGAUAGUUGGGUCUGUUGUUUUGUGGGCAACUUUUGGGUUAUUCUUCGCCCGUAGCUGUCGCUGACACUUGGCUGCCCACAUUUAUAUAUUUUUCUAUUCCGCGACC (((((((((((..((((.(((.....((((((((.((...((((........))))(((((....))).))....)).))))))))...))).))))..))).)))))))). ( -38.50) >DroSec_CAF1 18255 112 + 1 GUUGUGGAUAGUUGGGUCUGUUGUUUUGUGGCCAACUUUUGGGUUAUUCUUCGCCCGUAGCUGUCGCUGACACUUGGCUGCCCACAUUUAUAUAUUUUUCUAUUCCGCGACC (((((((((((..((((.((((((...(..(((((.....((((........))))(((((....))).))..)))))..)..)))...))).))))..))).)))))))). ( -33.80) >DroEre_CAF1 18487 112 + 1 GUUGUGGAUAGUUGGGUCUGUUGUUUUGUGGGCAACUUUUAGGUUAUUCUUUGCCCGUAGCUGUCGCUGACACUUGGCUGCCCACAUUUAUAUAUUUUUCUAUUCCGCGAGC .((((((((((..((((.(((.....(((((((((((....)))).......(((...((.((((...)))))).))).)))))))...))).))))..))).))))))).. ( -33.90) >DroYak_CAF1 47092 112 + 1 GUUGGGGAUAGUUGGGUCUGUUGUUUUGUGGGCAACUUUUAUGUUAUUCUUCGCCCUUAGCUGUCGCAGACACUUGGUUGCCCACUUUUAAAUAUUUUUCUUUUCCGAGACC (((.((((.((..((((...(((....((((((((((....((((.......((.....)).......))))...))))))))))...)))..))))..)).))))..))). ( -27.84) >consensus GUUGUGGAUAGUUGGGUCUGUUGUUUUGUGGGCAACUUUUAGGUUAUUCUUCGCCCGUAGCUGUCGCUGACACUUGGCUGCCCACAUUUAUAUAUUUUUCUAUUCCGCGACC (((((((((((..((((.(((.....((((((((((......))).......(((...((.((((...)))))).))).)))))))...))).))))..))).)))))))). (-28.14 = -29.70 + 1.56)


| Location | 15,826,143 – 15,826,255 |
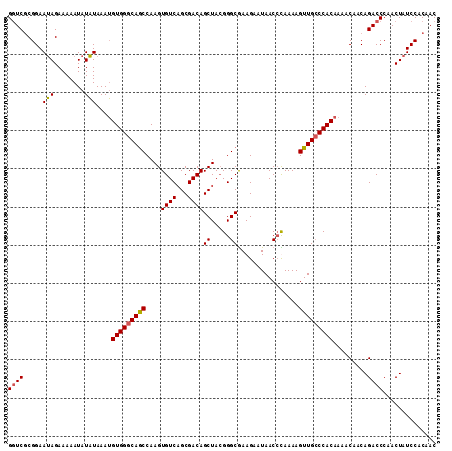
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 94.05 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -21.79 |
| Energy contribution | -21.72 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15826143 112 - 22407834 GGUCGCGGAAUAGAAAAAUAUAUAAAUGUGGGCAGCCAAGUGUCAGCGACAGCUACGGGCGAAGAAUAACCCAAAAGUUGCCCACAAAACAACAGACCCAACUAUCCACAAC ((((.....(((......))).....((((((((((.....((.(((....)))))(((..........)))....))))))))))........)))).............. ( -29.90) >DroSec_CAF1 18255 112 - 1 GGUCGCGGAAUAGAAAAAUAUAUAAAUGUGGGCAGCCAAGUGUCAGCGACAGCUACGGGCGAAGAAUAACCCAAAAGUUGGCCACAAAACAACAGACCCAACUAUCCACAAC ((((.....(((......))).....(((((.((((.....((.(((....)))))(((..........)))....)))).)))))........)))).............. ( -25.30) >DroEre_CAF1 18487 112 - 1 GCUCGCGGAAUAGAAAAAUAUAUAAAUGUGGGCAGCCAAGUGUCAGCGACAGCUACGGGCAAAGAAUAACCUAAAAGUUGCCCACAAAACAACAGACCCAACUAUCCACAAC ....(.(((.(((.............((((((((((.....((.(((....)))))((...........)).....))))))))))...(....)......)))))).)... ( -26.00) >DroYak_CAF1 47092 112 - 1 GGUCUCGGAAAAGAAAAAUAUUUAAAAGUGGGCAACCAAGUGUCUGCGACAGCUAAGGGCGAAGAAUAACAUAAAAGUUGCCCACAAAACAACAGACCCAACUAUCCCCAAC ((((((......)).............(((((((((....((((...))))((.....))................))))))))).........)))).............. ( -25.90) >consensus GGUCGCGGAAUAGAAAAAUAUAUAAAUGUGGGCAGCCAAGUGUCAGCGACAGCUACGGGCGAAGAAUAACCCAAAAGUUGCCCACAAAACAACAGACCCAACUAUCCACAAC ((((.....(((........)))....(((((((((....((((...))))((.....))................))))))))).........)))).............. (-21.79 = -21.72 + -0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:05 2006