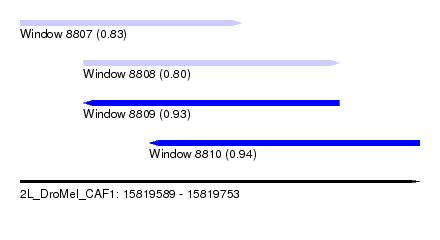
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,819,589 – 15,819,753 |
| Length | 164 |
| Max. P | 0.938808 |

| Location | 15,819,589 – 15,819,680 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 78.75 |
| Mean single sequence MFE | -19.78 |
| Consensus MFE | -8.30 |
| Energy contribution | -8.30 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15819589 91 + 22407834 CGAUUUAUCUACCGGAGUUGUAGGGAUCUAUCUCCCUAU--ACGAGUAGACGAGUAUGUAUUUAAUAUGAAUAUAU--AUAUACUCAUGUAUUGU .......(((((((....((((((((......)))))))--))).))))).((((((((((.((.......)).))--))))))))......... ( -25.90) >DroSec_CAF1 14143 84 + 1 CGAUUUAACUGCAGGAGGUGCAGAGAUCUAUCUCUCAAU--ACG--------AGUAUGUAUUUAAUAUGAAUAUAUAUAUAUACUCAUGUAUUG- .(((((..(((((.....)))))))))).......((((--(((--------(((((((((...(((....)))..))))))))))..))))))- ( -20.80) >DroSim_CAF1 14149 80 + 1 CGAUUUAUCAGCAGGAGGUGCAGAGAUCUAUCUCCCUAU--ACG--------AGUAUGUAUUUAAUAUGAAUAUA----UAUACUCAUGUAUUG- ..........((((((((((........)))))))....--..(--------(((((((((...........)))----))))))).)))....- ( -18.50) >DroEre_CAF1 13939 76 + 1 CGCUUUAUCUACAGGAGGCGCAGAGAUC----UCCCAAUAUGCG--------AAUAUGUGUUUAAUAUGAAUAU------AUACUCAUGUAUUG- .............((((((.....).))----)))((((((..(--------(.(((((((((.....))))))------))).))..))))))- ( -16.90) >DroYak_CAF1 40606 76 + 1 CGAUUGAUCUACAGGAGGUGCAGGGAUC----UCCUUAUAUGCG--------AAUAUGUGUUUAAUAUGAAUAU------AUACUCAUGUAUCG- (((.........(((((((......)))----))))((((((.(--------..(((((((((.....))))))------)))).)))))))))- ( -16.80) >consensus CGAUUUAUCUACAGGAGGUGCAGAGAUCUAUCUCCCUAU__ACG________AGUAUGUAUUUAAUAUGAAUAUA____UAUACUCAUGUAUUG_ .........((((.(((.....((((......)))).....................((((((.....)))))).........))).)))).... ( -8.30 = -8.30 + -0.00)

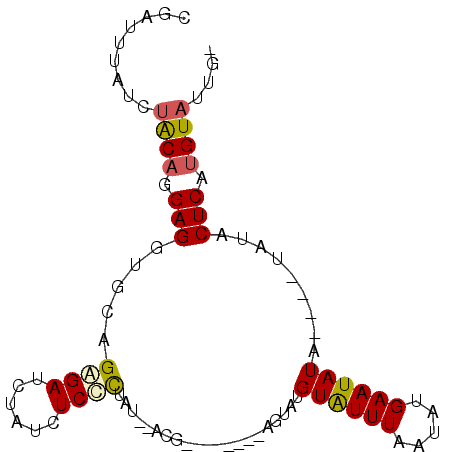
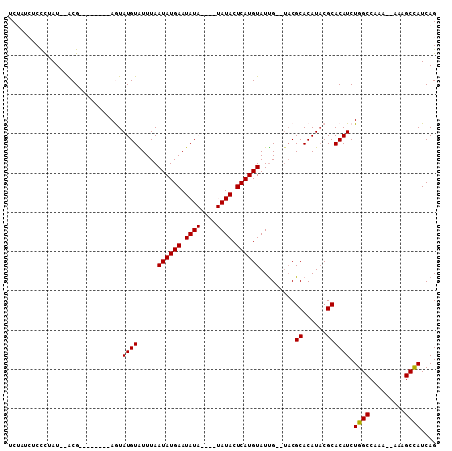
| Location | 15,819,615 – 15,819,720 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 83.00 |
| Mean single sequence MFE | -19.48 |
| Consensus MFE | -10.30 |
| Energy contribution | -10.24 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15819615 105 + 22407834 UCUAUCUCCCUAU--ACGAGUAGACGAGUAUGUAUUUAAUAUGAAUAUAU--AUAUACUCAUGUAUUGUGUACGCACAUACGCACAUCUGGCCAAAAAAAAGCCAUCAG ((((.(((.....--..))))))).((((((((((.((.......)).))--))))))))..((((.(((....))))))).......((((.........)))).... ( -22.90) >DroSec_CAF1 14169 95 + 1 UCUAUCUCUCAAU--ACG--------AGUAUGUAUUUAAUAUGAAUAUAUAUAUAUACUCAUGUAUUG--UACGCACAUACGCACAUCUGGCCAAA--AAAGCCAUCAG .........((((--(((--------(((((((((...(((....)))..))))))))))..))))))--...((......)).....((((....--...)))).... ( -19.10) >DroSim_CAF1 14175 91 + 1 UCUAUCUCCCUAU--ACG--------AGUAUGUAUUUAAUAUGAAUAUA----UAUACUCAUGUAUUG--UACGCACAUACGCACAUCUGGCCAAA--AAAGCCAUCAG ..........(((--(.(--------(((((((((...........)))----))))))).)))).((--(.((......)).)))..((((....--...)))).... ( -16.60) >DroEre_CAF1 13965 87 + 1 UC----UCCCAAUAUGCG--------AAUAUGUGUUUAAUAUGAAUAU------AUACUCAUGUAUUG--UUCGCACAUUCGCACAUCUGGCCAAA--AAAGCCAUCAG ..----........((((--------(((.((((..((((((((....------....))))))..))--..)))).)))))))....((((....--...)))).... ( -21.10) >DroYak_CAF1 40632 87 + 1 UC----UCCUUAUAUGCG--------AAUAUGUGUUUAAUAUGAAUAU------AUACUCAUGUAUCG--AACGCACAUACGCACAUCUGGCCAAA--AAAGCUAUCAG ..----.((..((.((((--------.((.(((((((.((((((....------....))))))...)--)))))).)).)))).))..)).....--........... ( -17.70) >consensus UCUAUCUCCCUAU__ACG________AGUAUGUAUUUAAUAUGAAUAUA____UAUACUCAUGUAUUG__UACGCACAUACGCACAUCUGGCCAAA__AAAGCCAUCAG .............................((((.....((((((.((((....)))).)))))).........((......)))))).((((.........)))).... (-10.30 = -10.24 + -0.06)

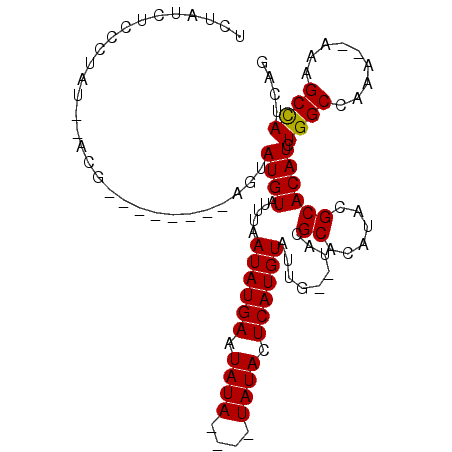
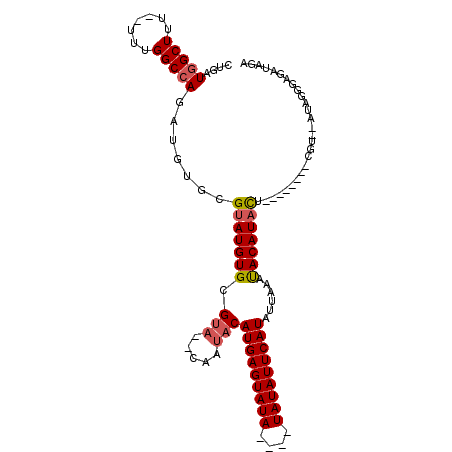
| Location | 15,819,615 – 15,819,720 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 83.00 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -15.94 |
| Energy contribution | -16.36 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15819615 105 - 22407834 CUGAUGGCUUUUUUUUGGCCAGAUGUGCGUAUGUGCGUACACAAUACAUGAGUAUAU--AUAUAUUCAUAUUAAAUACAUACUCGUCUACUCGU--AUAGGGAGAUAGA ....(((((.......)))))..(((((((....)))))))........((((((.(--((.((.......)).))).))))))((((.(((..--...)))))))... ( -25.80) >DroSec_CAF1 14169 95 - 1 CUGAUGGCUUU--UUUGGCCAGAUGUGCGUAUGUGCGUA--CAAUACAUGAGUAUAUAUAUAUAUUCAUAUUAAAUACAUACU--------CGU--AUUGAGAGAUAGA ....(((((..--...)))))..(((((((....)))))--))....(((((((((....)))))))))............((--------(..--.....)))..... ( -23.80) >DroSim_CAF1 14175 91 - 1 CUGAUGGCUUU--UUUGGCCAGAUGUGCGUAUGUGCGUA--CAAUACAUGAGUAUA----UAUAUUCAUAUUAAAUACAUACU--------CGU--AUAGGGAGAUAGA ....(((((..--...)))))..(((((((....)))))--))....((((((((.----(((...........))).)))))--------)))--............. ( -21.90) >DroEre_CAF1 13965 87 - 1 CUGAUGGCUUU--UUUGGCCAGAUGUGCGAAUGUGCGAA--CAAUACAUGAGUAU------AUAUUCAUAUUAAACACAUAUU--------CGCAUAUUGGGA----GA ....(((((..--...)))))((((((((((((((.(..--.((((..(((((..------..)))))))))...).))))))--------))))))))....----.. ( -27.40) >DroYak_CAF1 40632 87 - 1 CUGAUAGCUUU--UUUGGCCAGAUGUGCGUAUGUGCGUU--CGAUACAUGAGUAU------AUAUUCAUAUUAAACACAUAUU--------CGCAUAUAAGGA----GA ...........--.....((..(((((((.(((((.(((--..(((..(((((..------..))))))))..))).))))).--------)))))))..)).----.. ( -23.50) >consensus CUGAUGGCUUU__UUUGGCCAGAUGUGCGUAUGUGCGUA__CAAUACAUGAGUAUA____UAUAUUCAUAUUAAAUACAUACU________CGU__AUAGGGAGAUAGA ....(((((.......))))).......(((((((.(((.....)))(((((((((....)))))))))......)))))))........................... (-15.94 = -16.36 + 0.42)


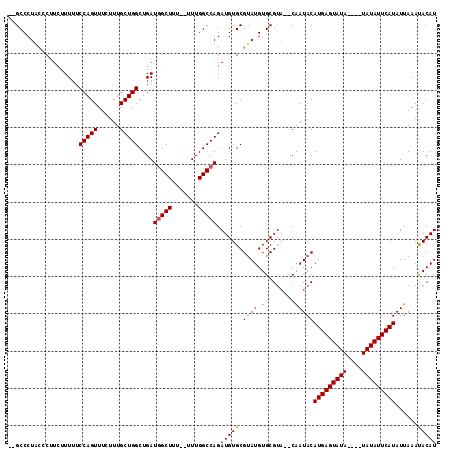
| Location | 15,819,642 – 15,819,753 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 89.85 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -20.02 |
| Energy contribution | -20.12 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15819642 111 - 22407834 --GCCCUACCCUUCUUUUUCCAGUUUCUUUGCUGGCUGAUGGCUUUUUUUUGGCCAGAUGUGCGUAUGUGCGUACACAAUACAUGAGUAUAU--AUAUAUUCAUAUUAAAUACAU --.................(((((......)))))....(((((.......)))))..(((((((....)))))))......((((((((..--..))))))))........... ( -25.60) >DroSec_CAF1 14188 109 - 1 --GCCCUACCCUUCUUUUUCCAGUUUCUUUGCUGGCUGAUGGCUUU--UUUGGCCAGAUGUGCGUAUGUGCGUA--CAAUACAUGAGUAUAUAUAUAUAUUCAUAUUAAAUACAU --.................(((((......)))))....(((((..--...)))))..(((((((....)))))--))....(((((((((....)))))))))........... ( -27.30) >DroSim_CAF1 14194 105 - 1 --GCCCUACCCUUCUUUUUCCAGUUUCUUUGCUGGCUGAUGGCUUU--UUUGGCCAGAUGUGCGUAUGUGCGUA--CAAUACAUGAGUAUA----UAUAUUCAUAUUAAAUACAU --.................(((((......)))))....(((((..--...)))))..(((((((....)))))--))....(((((((..----..)))))))........... ( -24.90) >DroEre_CAF1 13982 105 - 1 UCGCCCUUCGCCUGCUUUUCCAGUUUCUUUGCUGGCUGAUGGCUUU--UUUGGCCAGAUGUGCGAAUGUGCGAA--CAAUACAUGAGUAU------AUAUUCAUAUUAAACACAU (((((.(((((........(((((......)))))....(((((..--...))))).....))))).).)))).--......((((((..------..))))))........... ( -27.80) >DroYak_CAF1 40649 103 - 1 --GCCCUUUUCUUCUUUUUCCAGUUUCUUUGCUGGCUGAUAGCUUU--UUUGGCCAGAUGUGCGUAUGUGCGUU--CGAUACAUGAGUAU------AUAUUCAUAUUAAACACAU --....................((((.....(((((..(.......--.)..)))))(((((.((((((((...--..........))))------)))).))))).)))).... ( -21.92) >consensus __GCCCUACCCUUCUUUUUCCAGUUUCUUUGCUGGCUGAUGGCUUU__UUUGGCCAGAUGUGCGUAUGUGCGUA__CAAUACAUGAGUAUA____UAUAUUCAUAUUAAAUACAU ...................(((((......)))))....(((((.......))))).(((((.((((.((......))))))(((((((((....)))))))))......))))) (-20.02 = -20.12 + 0.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:59 2006