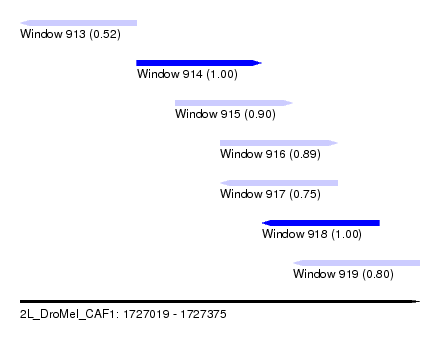
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,727,019 – 1,727,375 |
| Length | 356 |
| Max. P | 0.999292 |

| Location | 1,727,019 – 1,727,123 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -14.18 |
| Energy contribution | -14.26 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1727019 104 - 22407834 ACCGUGGAGAAAGUCACACCCGAAUGGGGAUAUAUGUAUAAGUUUACACCACUCACAGCUCAAAUUGCAAGUGACUUACAUGCUCACAGCUACAAUUCUUUGCA ...(..(((((((((((.(((.....))).....((((......)))).........((.......))..)))))))....((.....))......))))..). ( -24.80) >DroSec_CAF1 9154 104 - 1 UCCGCGGAGAAAGUCACACCCGAAUGGGGAUAUAUGUAUAACUUUACACCACUUACCGCUCAAAUUGCAAGUGACUUUCAUGCUCACAGCUAUAGUUCUUUGCA ...((((((((((((((.(((.....))).....((((......)))).........((.......))..)))))))))..((.....))........))))). ( -26.40) >DroSim_CAF1 9359 95 - 1 UCCGUGGAGAAAGUCACACCCGAAUGGGGAUAUAUGUAUAACUUUACACCACUCACCGCUCAAAUUGCAAGU---------GCUCACAGCUACAAUUCUUUGCA ...(..(((((..........((.(((.((....((((......))))....)).))).)).........((---------((.....)).))..)))))..). ( -17.50) >DroEre_CAF1 9169 95 - 1 UCCGUGGAGAAAGUCACACCCAAGUGGGGAUACA-----AAACUUACACCACUCGCCGCUCAAAUUGCAAGUGA----CAUGCUCACGCCUACAAUUCUCUGCA ...(..(((((.(((((.....(((((.......-----.........)))))....((.......))..))))----)..((....))......)))))..). ( -24.79) >DroYak_CAF1 9941 85 - 1 UCCGUGGAGAAAGUCACACCCAAGUGGGGAUACA----UAAACUUACACCACUCGCCACUCAAAUUGCAAGUGA---------------UUACAAUUCUUUGCA ...(..(((((((((((...(((((((.((....----..............)).))))).....))...))))---------------))....)))))..). ( -18.77) >consensus UCCGUGGAGAAAGUCACACCCGAAUGGGGAUAUAUGUAUAAAUUUACACCACUCACCGCUCAAAUUGCAAGUGA____CAUGCUCACAGCUACAAUUCUUUGCA ...((((((((.......(((....))).....................((((....((.......)).))))......................)))))))). (-14.18 = -14.26 + 0.08)

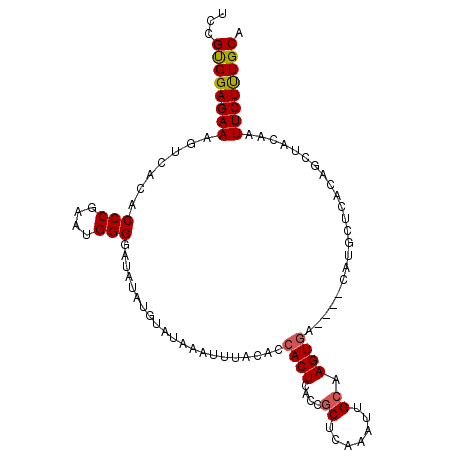
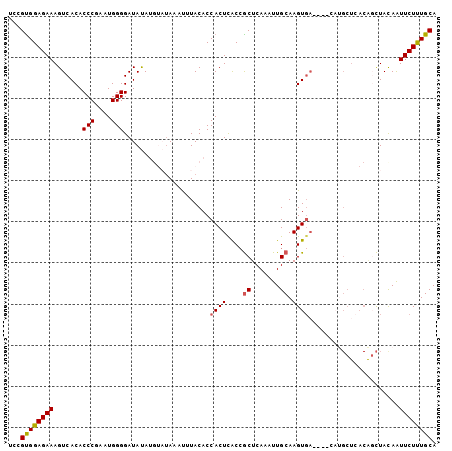
| Location | 1,727,123 – 1,727,234 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.82 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -22.00 |
| Energy contribution | -22.28 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.49 |
| SVM RNA-class probability | 0.999292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1727123 111 + 22407834 AUA------AUCUCGAAAUGCCCCUCCUGAGUUUUCUAUAUUUAAUUUGUCUUCAAAUUUCAGUUAAAUAUUUCUAUAAUAAUUUAGAGUCACUGACACUGGCGGCAUUUCCCGCUU--- ...------.....((((((((((...((..............((((((....))))))(((((.......(((((........)))))..)))))))..)).))))))))......--- ( -21.70) >DroSec_CAF1 9258 111 + 1 AUA------AUGUCGAAACGCCCCUCCUGCGUUUUCUAUAUUUAAUUUGUCUUCAGAUUUCAGUUAAAUAUUUCUAUAAUUAUUUAGAGUCACUGACACGGGCGGCAUUUCCCGCUU--- ...------.(((((((((((.......)))))))..((((((((((.(((....)))...))))))))))(((((........))))).....)))).((((((......))))))--- ( -24.40) >DroSim_CAF1 9454 111 + 1 AUA------AUGUCGAAAUGCCCCUCCUGCGUUUUCUAUAUUUAAUUUGUCUUCAGAUUUCAGUUAAAUAUUUCUAUAAUUAUUUAGAGUCACUGACACGGGCGGCAUUUCCCGCUU--- ...------.....((((((((...((((.(((....((((((((((.(((....)))...))))))))))(((((........))))).....))).)))).))))))))......--- ( -26.40) >DroEre_CAF1 9264 114 + 1 AUA------GUCUCGAAAUGCCCCCACUGCGAUUUCUAUAUUUAAUUUGUCUUCAGAUUUCAGUUAAAUAUUUCUAAAAUUAUUUGGAGUCACUGACACGGGCGGCAUUUCCCGCUUUGC ..(------((...(((((((((((............((((((((((.(((....)))...))))))))))(((((((....)))))))..........))).))))))))..))).... ( -27.70) >DroYak_CAF1 10026 120 + 1 AUACUCGUAUUCUCGAAAUGCCCCCUCUGCGUUUUCUAUAUUUAAUUUGUCUUAAGAUUUCAGUUAAAUAUUUCUAAAAUUAUUUAGAGUCACUGACACGGGCGGCAUUUCCCGCUUUGC ..............(((((((((((............((((((((((.(((....)))...))))))))))(((((((....)))))))..........))).))))))))......... ( -27.20) >consensus AUA______AUCUCGAAAUGCCCCUCCUGCGUUUUCUAUAUUUAAUUUGUCUUCAGAUUUCAGUUAAAUAUUUCUAUAAUUAUUUAGAGUCACUGACACGGGCGGCAUUUCCCGCUU___ ..............(((((((((((............((((((((((.(((....)))...))))))))))(((((........)))))..........))).))))))))......... (-22.00 = -22.28 + 0.28)

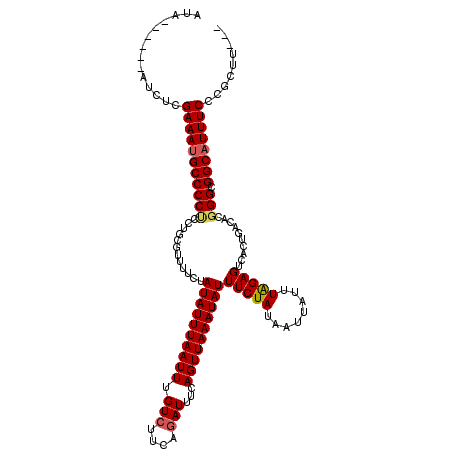
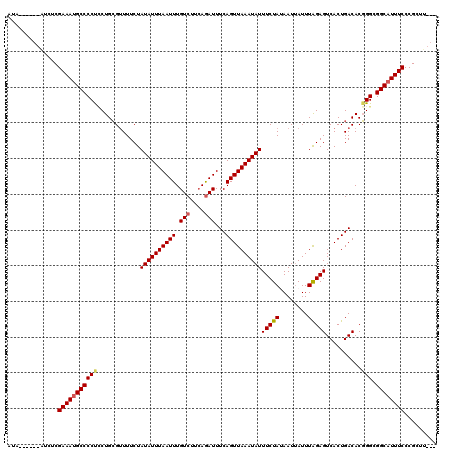
| Location | 1,727,157 – 1,727,262 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.25 |
| Mean single sequence MFE | -26.16 |
| Consensus MFE | -19.88 |
| Energy contribution | -19.96 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1727157 105 + 22407834 UUUAAUUUGUCUUCAAAUUUCAGUUAAAUAUUUCUAUAAUAAUUUAGAGUCACUGACACUGGCGGCAUUUCCCGCUU--------UUCCGCUUAUCUCAGUGGGAGAGUUUUC------- .......((((.(((....(((((.......(((((........)))))..)))))...))).))))......((((--------(((((((......)))))))))))....------- ( -23.10) >DroSec_CAF1 9292 104 + 1 UUUAAUUUGUCUUCAGAUUUCAGUUAAAUAUUUCUAUAAUUAUUUAGAGUCACUGACACGGGCGGCAUUUCCCGCUU--------UUCCGCUU-UCUCAGUGGGAGAGUUUUC------- ...((((((....))))))(((((.......(((((........)))))..)))))...(((........)))((((--------(((((((.-....)))))))))))....------- ( -23.50) >DroSim_CAF1 9488 104 + 1 UUUAAUUUGUCUUCAGAUUUCAGUUAAAUAUUUCUAUAAUUAUUUAGAGUCACUGACACGGGCGGCAUUUCCCGCUU--------UUCCGCUU-UCUCAGUGGGAGAGUUUUC------- ...((((((....))))))(((((.......(((((........)))))..)))))...(((........)))((((--------(((((((.-....)))))))))))....------- ( -23.50) >DroEre_CAF1 9298 112 + 1 UUUAAUUUGUCUUCAGAUUUCAGUUAAAUAUUUCUAAAAUUAUUUGGAGUCACUGACACGGGCGGCAUUUCCCGCUUUGCCGCUUUUCCGCUG-UCUCAGUGGGAGAGUUUUC------- .......((((....((((((((.(((............))).))))))))...)))).((((((......))))))....((((((((((((-...))))))))))))....------- ( -30.70) >DroYak_CAF1 10066 119 + 1 UUUAAUUUGUCUUAAGAUUUCAGUUAAAUAUUUCUAAAAUUAUUUAGAGUCACUGACACGGGCGGCAUUUCCCGCUUUGCCGCUUUUCCGCUU-UCUCAGUGGGAGAGUUUUCAGUUUUC (((((((.(((....)))...)))))))...(((((((....)))))))..(((((...((((((......))))))....(((((((((((.-....)))))))))))..))))).... ( -30.00) >consensus UUUAAUUUGUCUUCAGAUUUCAGUUAAAUAUUUCUAUAAUUAUUUAGAGUCACUGACACGGGCGGCAUUUCCCGCUU________UUCCGCUU_UCUCAGUGGGAGAGUUUUC_______ ...((((((....))))))(((((.......(((((........)))))..)))))...((((((......))))))........(((((((......)))))))............... (-19.88 = -19.96 + 0.08)

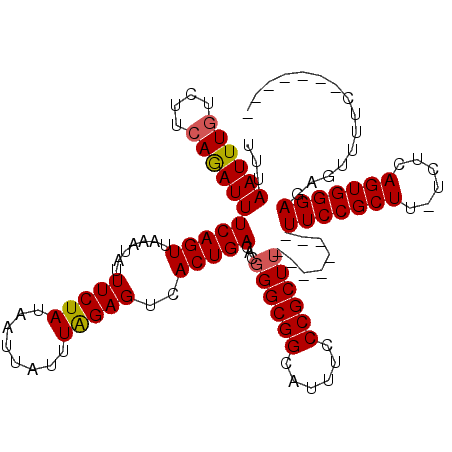
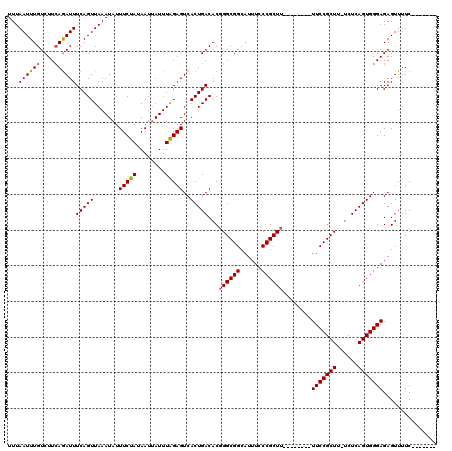
| Location | 1,727,197 – 1,727,302 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.25 |
| Mean single sequence MFE | -32.65 |
| Consensus MFE | -25.58 |
| Energy contribution | -25.82 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.887105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1727197 105 + 22407834 AAUUUAGAGUCACUGACACUGGCGGCAUUUCCCGCUU--------UUCCGCUUAUCUCAGUGGGAGAGUUUUC-------AGUUUGCUCUCGCCGAACAACCGAAUGACCUCUAAGCUAC ..((((((((((..........((((...........--------..(((((......)))))((((((....-------.....))))))))))..........))).))))))).... ( -29.25) >DroSec_CAF1 9332 104 + 1 UAUUUAGAGUCACUGACACGGGCGGCAUUUCCCGCUU--------UUCCGCUU-UCUCAGUGGGAGAGUUUUC-------AGUUUGCUCUCGCCGAACAACCGAAUGACCUCUAAGCUAC ..((((((((((......(((.((((...........--------..(((((.-....)))))((((((....-------.....)))))))))).....)))..))).))))))).... ( -30.80) >DroSim_CAF1 9528 104 + 1 UAUUUAGAGUCACUGACACGGGCGGCAUUUCCCGCUU--------UUCCGCUU-UCUCAGUGGGAGAGUUUUC-------AGUUUGCUCUCGCCGAACAACCGAAUGACCUCUAAGCUAC ..((((((((((......(((.((((...........--------..(((((.-....)))))((((((....-------.....)))))))))).....)))..))).))))))).... ( -30.80) >DroEre_CAF1 9338 112 + 1 UAUUUGGAGUCACUGACACGGGCGGCAUUUCCCGCUUUGCCGCUUUUCCGCUG-UCUCAGUGGGAGAGUUUUC-------AGUUUUCUCUCGCCGAACAACCGAAUGACCUCUAAGCUAC ..((((((((((..(((((((((((((..........))))))....))).))-))((.(((((((((.....-------....))))))))).)).........))).))))))).... ( -37.70) >DroYak_CAF1 10106 119 + 1 UAUUUAGAGUCACUGACACGGGCGGCAUUUCCCGCUUUGCCGCUUUUCCGCUU-UCUCAGUGGGAGAGUUUUCAGUUUUCAGUUUUCUCUCUGCGAACAACCGAAUGACCUCUAAGCUAC ..((((((((((......(((((((((..........))))))(((((((((.-....))))))))).......((((.(((........))).))))..)))..))).))))))).... ( -34.70) >consensus UAUUUAGAGUCACUGACACGGGCGGCAUUUCCCGCUU________UUCCGCUU_UCUCAGUGGGAGAGUUUUC_______AGUUUGCUCUCGCCGAACAACCGAAUGACCUCUAAGCUAC ..((((((((((((((...((((((......................))))))...)))))))(((((..................)))))..................))))))).... (-25.58 = -25.82 + 0.24)

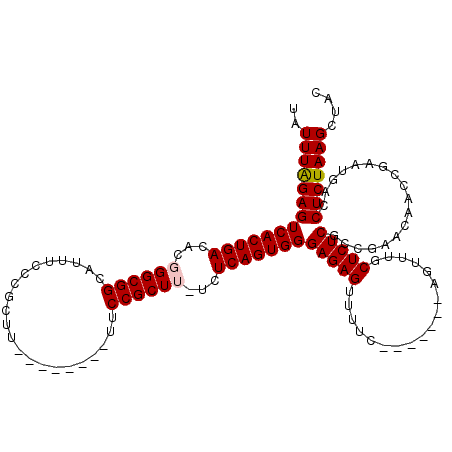
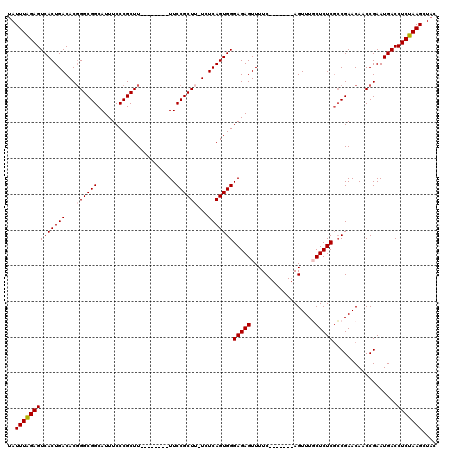
| Location | 1,727,197 – 1,727,302 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.25 |
| Mean single sequence MFE | -35.07 |
| Consensus MFE | -25.53 |
| Energy contribution | -26.13 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1727197 105 - 22407834 GUAGCUUAGAGGUCAUUCGGUUGUUCGGCGAGAGCAAACU-------GAAAACUCUCCCACUGAGAUAAGCGGAA--------AAGCGGGAAAUGCCGCCAGUGUCAGUGACUCUAAAUU .....((((((....(((((((((((.....)))).))))-------)))........((((((.((..((((..--------............))))..)).)))))).))))))... ( -33.04) >DroSec_CAF1 9332 104 - 1 GUAGCUUAGAGGUCAUUCGGUUGUUCGGCGAGAGCAAACU-------GAAAACUCUCCCACUGAGA-AAGCGGAA--------AAGCGGGAAAUGCCGCCCGUGUCAGUGACUCUAAAUA .....((((((....(((((((((((.....)))).))))-------)))........((((((..-..((((..--------..((((......)))))))).)))))).))))))... ( -35.70) >DroSim_CAF1 9528 104 - 1 GUAGCUUAGAGGUCAUUCGGUUGUUCGGCGAGAGCAAACU-------GAAAACUCUCCCACUGAGA-AAGCGGAA--------AAGCGGGAAAUGCCGCCCGUGUCAGUGACUCUAAAUA .....((((((....(((((((((((.....)))).))))-------)))........((((((..-..((((..--------..((((......)))))))).)))))).))))))... ( -35.70) >DroEre_CAF1 9338 112 - 1 GUAGCUUAGAGGUCAUUCGGUUGUUCGGCGAGAGAAAACU-------GAAAACUCUCCCACUGAGA-CAGCGGAAAAGCGGCAAAGCGGGAAAUGCCGCCCGUGUCAGUGACUCCAAAUA ........(..((((((((((.....((.(((((......-------.....))))))))))))((-((.(((....((((((..........))))))))))))).)))))..)..... ( -34.80) >DroYak_CAF1 10106 119 - 1 GUAGCUUAGAGGUCAUUCGGUUGUUCGCAGAGAGAAAACUGAAAACUGAAAACUCUCCCACUGAGA-AAGCGGAAAAGCGGCAAAGCGGGAAAUGCCGCCCGUGUCAGUGACUCUAAAUA .....((((((....(((((((.....(((........)))..)))))))........((((((..-..((((....((((((..........)))))))))).)))))).))))))... ( -36.10) >consensus GUAGCUUAGAGGUCAUUCGGUUGUUCGGCGAGAGCAAACU_______GAAAACUCUCCCACUGAGA_AAGCGGAA________AAGCGGGAAAUGCCGCCCGUGUCAGUGACUCUAAAUA .....((((((.((((((.(((.(((((.(((((..................)))))...)))))...))).))...........(((((........)))))....))))))))))... (-25.53 = -26.13 + 0.60)

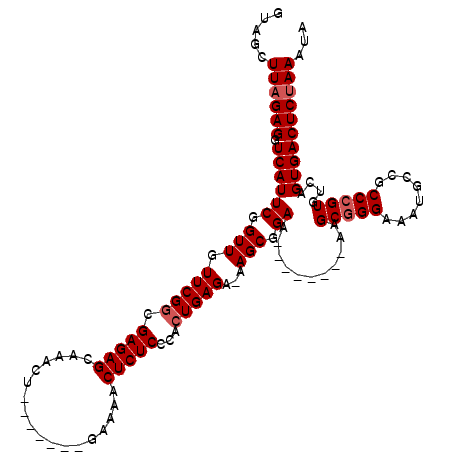
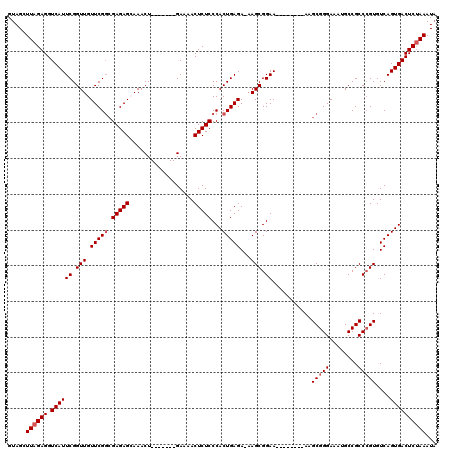
| Location | 1,727,234 – 1,727,339 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 91.33 |
| Mean single sequence MFE | -35.44 |
| Consensus MFE | -30.77 |
| Energy contribution | -30.33 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1727234 105 - 22407834 UUCGCUGGACGCCUCACUUUGGGUCUGACCUUUAAAAGUAGCUUAGAGGUCAUUCGGUUGUUCGGCGAGAGCAAACU-------GAAAACUCUCCCACUGAGAUAAGCGGAA----- ((((((......((((...((((..((((((((((.......))))))))))(((((((((((.....)))).))))-------)))......)))).))))...)))))).----- ( -36.70) >DroSec_CAF1 9369 104 - 1 UUCGCUGGACGCCUCACUUUGAGUCUGACCUUUAAAAGUAGCUUAGAGGUCAUUCGGUUGUUCGGCGAGAGCAAACU-------GAAAACUCUCCCACUGAGA-AAGCGGAA----- ((((((......((((....((((.((((((((((.......))))))))))(((((((((((.....)))).))))-------))).))))......)))).-.)))))).----- ( -34.10) >DroSim_CAF1 9565 104 - 1 UUCGCUGGACGCCUCACUUUGAGUCUGACCUUUAAAAGUAGCUUAGAGGUCAUUCGGUUGUUCGGCGAGAGCAAACU-------GAAAACUCUCCCACUGAGA-AAGCGGAA----- ((((((......((((....((((.((((((((((.......))))))))))(((((((((((.....)))).))))-------))).))))......)))).-.)))))).----- ( -34.10) >DroEre_CAF1 9378 109 - 1 UCUGCUGGACGCCUCACUUUGGGUCUGACCUUUAAAAGUAGCUUAGAGGUCAUUCGGUUGUUCGGCGAGAGAAAACU-------GAAAACUCUCCCACUGAGA-CAGCGGAAAAGCG ((((((((((.((.......)))))((((((((((.......))))))))))((((((.....((.(((((......-------.....))))))))))))).-)))))))...... ( -37.40) >DroYak_CAF1 10146 116 - 1 UUUGCUGGACGCCUCACUUUGGGUCUGACCUUUAAAAGUAGCUUAGAGGUCAUUCGGUUGUUCGCAGAGAGAAAACUGAAAACUGAAAACUCUCCCACUGAGA-AAGCGGAAAAGCG ...(((...(((((((...((((..((((((((((.......))))))))))(((((((.....(((........)))..)))))))......)))).)))).-..)))....))). ( -34.90) >consensus UUCGCUGGACGCCUCACUUUGGGUCUGACCUUUAAAAGUAGCUUAGAGGUCAUUCGGUUGUUCGGCGAGAGCAAACU_______GAAAACUCUCCCACUGAGA_AAGCGGAA_____ (((((((((((((((.....))(..((((((((((.......))))))))))..))).)))))((.(((((..................))))))).........))))))...... (-30.77 = -30.33 + -0.44)

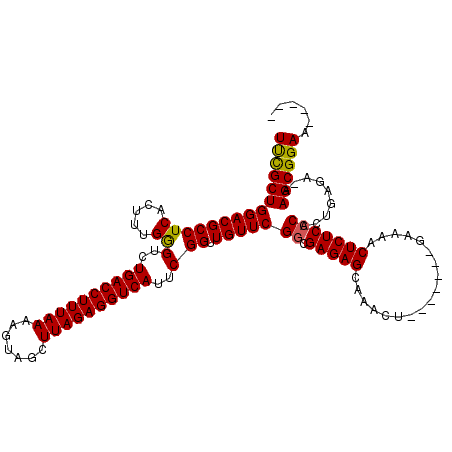
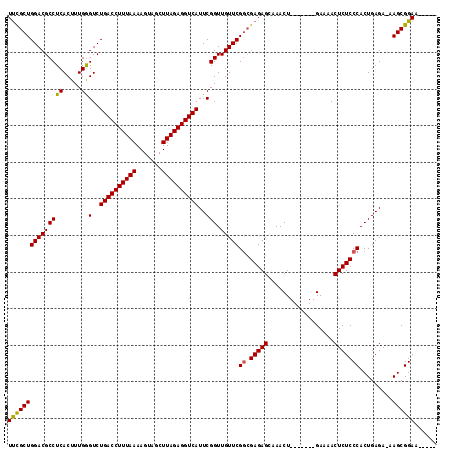
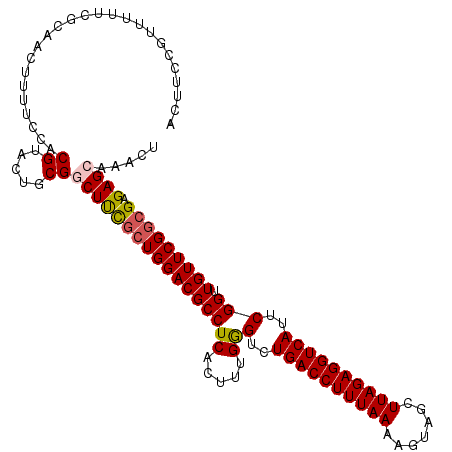
| Location | 1,727,262 – 1,727,375 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 94.51 |
| Mean single sequence MFE | -34.03 |
| Consensus MFE | -27.52 |
| Energy contribution | -27.68 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1727262 113 - 22407834 ACUUCCGUUUUUCGCAACUUUUCCACGUACUGCGGCUUCGCUGGACGCCUCACUUUGGGUCUGACCUUUAAAAGUAGCUUAGAGGUCAUUCGGUUGUUCGGCGAGAGCAAACU ......((((..((((((........))..))))((((((((((((((((......)))).((((((((((.......)))))))))).......))))))))).))))))). ( -35.80) >DroSec_CAF1 9396 113 - 1 ACUUCCGUUUUUUGCAACUUUUCCACGUACUUCGGCUUCGCUGGACGCCUCACUUUGAGUCUGACCUUUAAAAGUAGCUUAGAGGUCAUUCGGUUGUUCGGCGAGAGCAAACU ..........(((((..........((.....))..(((((((((((((((.....))(..((((((((((.......))))))))))..))).))))))))))).))))).. ( -33.80) >DroSim_CAF1 9592 113 - 1 ACUUCCGUUUUUUGCAACUUUUCCACGUACUUCGGCUUCGCUGGACGCCUCACUUUGAGUCUGACCUUUAAAAGUAGCUUAGAGGUCAUUCGGUUGUUCGGCGAGAGCAAACU ..........(((((..........((.....))..(((((((((((((((.....))(..((((((((((.......))))))))))..))).))))))))))).))))).. ( -33.80) >DroEre_CAF1 9410 113 - 1 ACUUUCCUUUUUCGCAACUUUUCCACGUCGUGCGGCUCUGCUGGACGCCUCACUUUGGGUCUGACCUUUAAAAGUAGCUUAGAGGUCAUUCGGUUGUUCGGCGAGAGAAAACU .......((((((((.......((.((..(((.(((((.....)).))).)))..)).(..((((((((((.......))))))))))..))).......))))))))..... ( -33.84) >DroYak_CAF1 10185 113 - 1 ACUUUCGUUUUUCGCAACUUUUCCACGUAGUGCGGCUUUGCUGGACGCCUCACUUUGGGUCUGACCUUUAAAAGUAGCUUAGAGGUCAUUCGGUUGUUCGCAGAGAGAAAACU .(((((.......((((((...(((...((((.(((..........))).)))).)))(..((((((((((.......))))))))))..))))))).....)))))...... ( -32.90) >consensus ACUUCCGUUUUUCGCAACUUUUCCACGUACUGCGGCUUCGCUGGACGCCUCACUUUGGGUCUGACCUUUAAAAGUAGCUUAGAGGUCAUUCGGUUGUUCGGCGAGAGCAAACU .........................((.....))(((((((((((((((((.....))(..((((((((((.......))))))))))..))).))))))))).))))..... (-27.52 = -27.68 + 0.16)

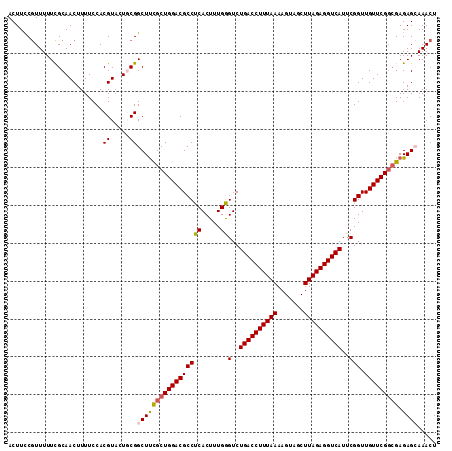
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:36 2006