| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,814,189 – 15,814,279 |
| Length | 90 |
| Max. P | 0.989408 |

| Location | 15,814,189 – 15,814,279 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -25.34 |
| Consensus MFE | -13.69 |
| Energy contribution | -15.92 |
| Covariance contribution | 2.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
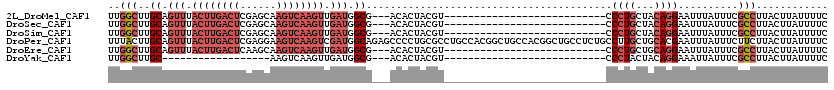
>2L_DroMel_CAF1 15814189 90 + 22407834 UUGGCUUGCAGUUUACUUGACUCGAGCAAGUCAAGUUGAUGGCG---ACACUACGU---------------------------CCCUGCUACAGGAAUUUAUUUCGCCUUACUUAUUUUC ..(((((((((((.....))))...)))))))((((....((((---(...((.((---------------------------.((((...)))).)).))..)))))..))))...... ( -24.20) >DroSec_CAF1 8862 90 + 1 UUGGCUUGCAGUUUACUUGACUCGAGCAAGUCAAGUUGAUGGCG---ACACUACGU---------------------------CCCUGCUACAGGAAUUUAUUUCGCCUUACUUAUUUUC ..(((((((((((.....))))...)))))))((((....((((---(...((.((---------------------------.((((...)))).)).))..)))))..))))...... ( -24.20) >DroSim_CAF1 8883 90 + 1 UUGGCUUGCAGUUUACUUGACUCGAGCAAGUCAAGUUGAUGGCG---ACACUACGU---------------------------CCCUGCUACAGGAAUUUAUUUCGCCUUACUUAUUUUC ..(((((((((((.....))))...)))))))((((....((((---(...((.((---------------------------.((((...)))).)).))..)))))..))))...... ( -24.20) >DroPer_CAF1 16661 120 + 1 UUUACUUGCAGUUUACUUGACUCGAGGAAGUCAAGUCGAUGGCAGAGCCCCUGCGCCUGCCACGGCUGCCACGGCUGCCUCUGCCUUGCUGCACGAAUUUAUUUCUUCUUACUUAUUUUC ....(.((((((..((((((((......))))))))....((((((((((.((.((..((....)).)))).))..).)))))))..)))))).)......................... ( -36.50) >DroEre_CAF1 8946 90 + 1 UUGGCUUGCAGUUUACUUGACUCAAGCAAGUCAAGUUGAUGGCG---ACACUACGU---------------------------CCCUGCUGCAGGAAUUUAUUUCGCCUUACUUAUUUUC ..((((((((((..((((((((......))))))))....((.(---((.....))---------------------------))).)))))))(((.....))))))............ ( -26.80) >DroYak_CAF1 34533 72 + 1 UUGGCUUGC------------------AAGUCAAGUUGAUGGCG---ACACUACGU---------------------------CCCUACUACAGGAAAUUAUUUCGCCUUACUUAUUUUC (..(((((.------------------....)))))..).((((---(.......(---------------------------((........))).......)))))............ ( -16.14) >consensus UUGGCUUGCAGUUUACUUGACUCGAGCAAGUCAAGUUGAUGGCG___ACACUACGU___________________________CCCUGCUACAGGAAUUUAUUUCGCCUUACUUAUUUUC ..(((..((.(((.((((((((......)))))))).))).)).........................................((((...))))..........)))............ (-13.69 = -15.92 + 2.22)
| Location | 15,814,189 – 15,814,279 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -14.61 |
| Energy contribution | -16.20 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
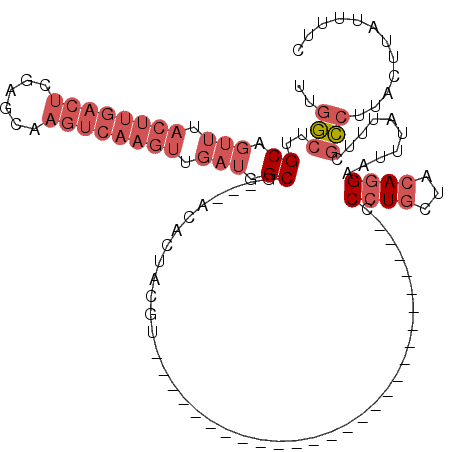
>2L_DroMel_CAF1 15814189 90 - 22407834 GAAAAUAAGUAAGGCGAAAUAAAUUCCUGUAGCAGGG---------------------------ACGUAGUGU---CGCCAUCAACUUGACUUGCUCGAGUCAAGUAAACUGCAAGCCAA ........(((.(((((..((..((((((...)))))---------------------------)..))...)---))))....(((((((((....)))))))))....)))....... ( -29.10) >DroSec_CAF1 8862 90 - 1 GAAAAUAAGUAAGGCGAAAUAAAUUCCUGUAGCAGGG---------------------------ACGUAGUGU---CGCCAUCAACUUGACUUGCUCGAGUCAAGUAAACUGCAAGCCAA ........(((.(((((..((..((((((...)))))---------------------------)..))...)---))))....(((((((((....)))))))))....)))....... ( -29.10) >DroSim_CAF1 8883 90 - 1 GAAAAUAAGUAAGGCGAAAUAAAUUCCUGUAGCAGGG---------------------------ACGUAGUGU---CGCCAUCAACUUGACUUGCUCGAGUCAAGUAAACUGCAAGCCAA ........(((.(((((..((..((((((...)))))---------------------------)..))...)---))))....(((((((((....)))))))))....)))....... ( -29.10) >DroPer_CAF1 16661 120 - 1 GAAAAUAAGUAAGAAGAAAUAAAUUCGUGCAGCAAGGCAGAGGCAGCCGUGGCAGCCGUGGCAGGCGCAGGGGCUCUGCCAUCGACUUGACUUCCUCGAGUCAAGUAAACUGCAAGUAAA ............(((........))).(((((...(((((((.(..((...((.(((......))))).))).)))))))....(((((((((....)))))))))...)))))...... ( -42.50) >DroEre_CAF1 8946 90 - 1 GAAAAUAAGUAAGGCGAAAUAAAUUCCUGCAGCAGGG---------------------------ACGUAGUGU---CGCCAUCAACUUGACUUGCUUGAGUCAAGUAAACUGCAAGCCAA ........(((.(((((..((..((((((...)))))---------------------------)..))...)---))))....(((((((((....)))))))))....)))....... ( -28.60) >DroYak_CAF1 34533 72 - 1 GAAAAUAAGUAAGGCGAAAUAAUUUCCUGUAGUAGGG---------------------------ACGUAGUGU---CGCCAUCAACUUGACUU------------------GCAAGCCAA ............(((((.((.((.((((......)))---------------------------).)).)).)---)))).....((((....------------------.)))).... ( -16.00) >consensus GAAAAUAAGUAAGGCGAAAUAAAUUCCUGUAGCAGGG___________________________ACGUAGUGU___CGCCAUCAACUUGACUUGCUCGAGUCAAGUAAACUGCAAGCCAA ............(((..........((((...)))).........................................((.....(((((((((....))))))))).....))..))).. (-14.61 = -16.20 + 1.59)

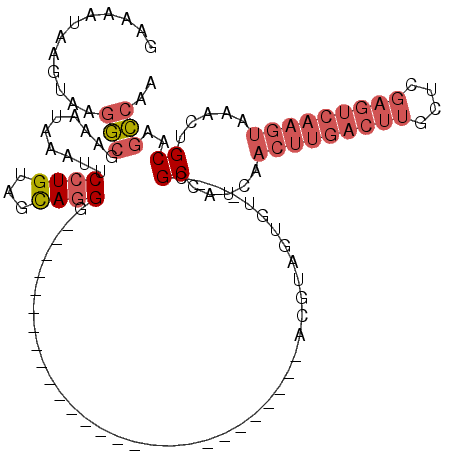
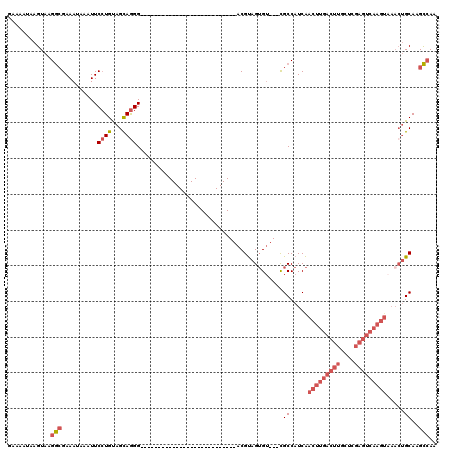
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:54 2006