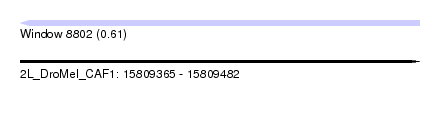
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,809,365 – 15,809,482 |
| Length | 117 |
| Max. P | 0.609929 |

| Location | 15,809,365 – 15,809,482 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.52 |
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -13.90 |
| Energy contribution | -14.82 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15809365 117 - 22407834 GUGCGUAACAAAUAGAAAUUUGCCAAGAAAAUAAUUGAACUUCAAUUGAGUGAGAGCAUGUGCAGAAUUUCAAGUAUACGUAUUACCGCCU---GUAUGUAUAGAAAGAGCCAAACGCAC ((((((........(((((((((((......((((((.....)))))).((....)).)).)))).)))))...(((((((((........---)))))))))...........)))))) ( -28.90) >DroSec_CAF1 3915 117 - 1 GUGCGUAACAAAUAGAAAUUUGCCAAGAAAAUAAUUGAACUUCAAUUGAGUGAGAGCAUGUGCAGAAUUUCAAGUAUACGUAUUACCGGCU---GUUCGUAUAGAAAGAGCCAAACGCAC ((((((........(((((((((((......((((((.....)))))).((....)).)).)))).)))))................((((---.(((.....)))..))))..)))))) ( -30.20) >DroSim_CAF1 3929 117 - 1 GUGCGUAACAAAUAGAAAUUUGCCAAGAAAAUAAUUGAACUUCAAUUGAGUGAGAGCAUGUGCAGAAUUUCAAGUAUACGUAUUACCGGCU---GUACGUAUAGGAAGAGCCAAACGCAC ((((((........(((((((((((......((((((.....)))))).((....)).)).)))).)))))...(((((((((........---)))))))))((.....))..)))))) ( -31.20) >DroPer_CAF1 8012 116 - 1 GUGCGUAGCCAAUAGAAAUUUGCUAGGAAAAUAAUUUAACUCCAAUUGAGUGCGAGCAUGUGCAGAAUUUCAAAUAA----AUUGCCGGAGCACACACUCGCAGGUUGAACAAAAAAUAA ((...(((((............................((((.....))))(((((..(((((..............----.........)))))..))))).))))).))......... ( -23.40) >DroEre_CAF1 4001 95 - 1 GUGCGUAACAAAUAGAAAUUUGCCAAGAAAAUAAUUGAACUUCAAUUGAGUGAGAGCAUGUGCAGAAUAUCAAGUAUACGCAUUACCGGCU---GUAU---------------------- (((((((((..(((....(((((((......((((((.....)))))).((....)).)).))))).)))...)).)))))))........---....---------------------- ( -17.70) >DroYak_CAF1 29004 95 - 1 GUGCGUAACAAAAAGAAAUUUGCCAAGAAAAUAAUUGAACUUCAAUUGAGUGAGAGCAUGUGCAGAAUUUCAAGUAUACGUAUUACCGACU---GUAU---------------------- (((((((((.....(((((((((((......((((((.....)))))).((....)).)).)))).)))))..)).)))))))........---....---------------------- ( -17.70) >consensus GUGCGUAACAAAUAGAAAUUUGCCAAGAAAAUAAUUGAACUUCAAUUGAGUGAGAGCAUGUGCAGAAUUUCAAGUAUACGUAUUACCGGCU___GUACGUAUAG_AAGAGCCAAACGCAC (((((((.......(((((((((((......((((((.....)))))).((....)).)).)))).))))).....)))))))..................................... (-13.90 = -14.82 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:52 2006