
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
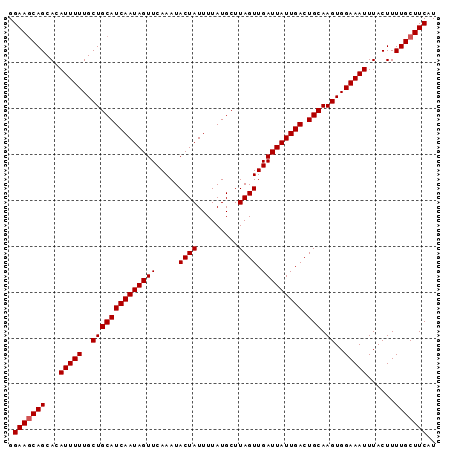
| Location | 15,809,161 – 15,809,293 |
| Length | 132 |
| Max. P | 0.999771 |

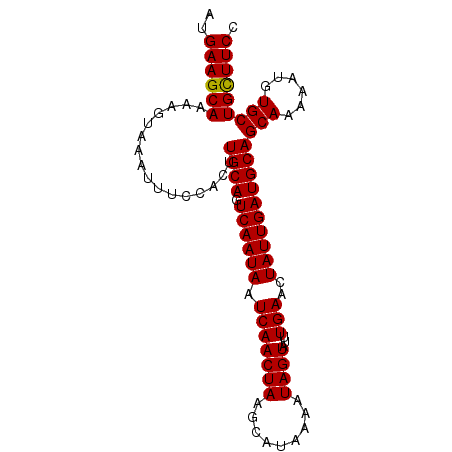
| Location | 15,809,161 – 15,809,254 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 99.57 |
| Mean single sequence MFE | -18.12 |
| Consensus MFE | -18.28 |
| Energy contribution | -18.12 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.08 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15809161 93 + 22407834 AUGAAGCAAAAGUAAAUUUCCACUUGCAGUCAAUAAUCAACUAAGCAUAAAAUAGUAUUUGAACUAUUGAUGCAGCAAAAAUGUGCUGCUUCC ..((((((................((((.((((((.(((((((.........))))...)))..))))))))))(((......))))))))). ( -18.60) >DroSec_CAF1 3711 93 + 1 AUGAAGCAAAAGUAAAUUUCCACUUGCAGUCAAUAAUCAACUAAGCAUAAAAUAGUAUUUGAACUAUUGAUGCAGCAAAAAUGUGCUGCUUCC ..((((((................((((.((((((.(((((((.........))))...)))..))))))))))(((......))))))))). ( -18.60) >DroSim_CAF1 3725 93 + 1 AUGAAGCAAAAGUAAAUUUCCACUUGCAGUCAAUAAUCAACUAAGCAUAAAAUAGUAUUUGAACUAUUGAUGCAGCAAAAAUGUGCUGCUUCC ..((((((................((((.((((((.(((((((.........))))...)))..))))))))))(((......))))))))). ( -18.60) >DroEre_CAF1 3801 93 + 1 AUGAAGCAAAAGUAAAUUUCCACUUGCAGUCAAUAAUCAACUAAGCAUAAAAUAGUAUUUGAACUAUUGAUGCAGCAAAAAUGUGCUGCUUCC ..((((((................((((.((((((.(((((((.........))))...)))..))))))))))(((......))))))))). ( -18.60) >DroYak_CAF1 28805 93 + 1 AUGAAGCAAAAGUAAAUUUCCACUUGCAGUCAAUAAUCAACUAAGCAUAAAAUAGUAUUUGAACUAUUGAUGCAGCAAAAAUGUGCUGUUUCC ..((((((................((((.((((((.(((((((.........))))...)))..))))))))))(((......))))))))). ( -16.20) >consensus AUGAAGCAAAAGUAAAUUUCCACUUGCAGUCAAUAAUCAACUAAGCAUAAAAUAGUAUUUGAACUAUUGAUGCAGCAAAAAUGUGCUGCUUCC ..((((((................((((.((((((.(((((((.........))))...)))..))))))))))(((......))))))))). (-18.28 = -18.12 + -0.16)

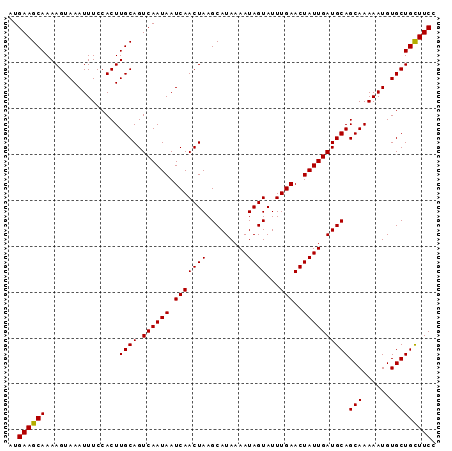
| Location | 15,809,161 – 15,809,254 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 99.57 |
| Mean single sequence MFE | -24.78 |
| Consensus MFE | -24.60 |
| Energy contribution | -24.80 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.04 |
| SVM RNA-class probability | 0.999771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15809161 93 - 22407834 GGAAGCAGCACAUUUUUGCUGCAUCAAUAGUUCAAAUACUAUUUUAUGCUUAGUUGAUUAUUGACUGCAAGUGGAAAUUUACUUUUGCUUCAU .(((((((...(((((..((((((((((((((.....((((.........)))).))))))))).))).))..)))))......))))))).. ( -25.90) >DroSec_CAF1 3711 93 - 1 GGAAGCAGCACAUUUUUGCUGCAUCAAUAGUUCAAAUACUAUUUUAUGCUUAGUUGAUUAUUGACUGCAAGUGGAAAUUUACUUUUGCUUCAU .(((((((...(((((..((((((((((((((.....((((.........)))).))))))))).))).))..)))))......))))))).. ( -25.90) >DroSim_CAF1 3725 93 - 1 GGAAGCAGCACAUUUUUGCUGCAUCAAUAGUUCAAAUACUAUUUUAUGCUUAGUUGAUUAUUGACUGCAAGUGGAAAUUUACUUUUGCUUCAU .(((((((...(((((..((((((((((((((.....((((.........)))).))))))))).))).))..)))))......))))))).. ( -25.90) >DroEre_CAF1 3801 93 - 1 GGAAGCAGCACAUUUUUGCUGCAUCAAUAGUUCAAAUACUAUUUUAUGCUUAGUUGAUUAUUGACUGCAAGUGGAAAUUUACUUUUGCUUCAU .(((((((...(((((..((((((((((((((.....((((.........)))).))))))))).))).))..)))))......))))))).. ( -25.90) >DroYak_CAF1 28805 93 - 1 GGAAACAGCACAUUUUUGCUGCAUCAAUAGUUCAAAUACUAUUUUAUGCUUAGUUGAUUAUUGACUGCAAGUGGAAAUUUACUUUUGCUUCAU (....)((((.(((((..((((((((((((((.....((((.........)))).))))))))).))).))..))))).......)))).... ( -20.30) >consensus GGAAGCAGCACAUUUUUGCUGCAUCAAUAGUUCAAAUACUAUUUUAUGCUUAGUUGAUUAUUGACUGCAAGUGGAAAUUUACUUUUGCUUCAU .(((((((...(((((..((((((((((((((.....((((.........)))).))))))))).))).))..)))))......))))))).. (-24.60 = -24.80 + 0.20)


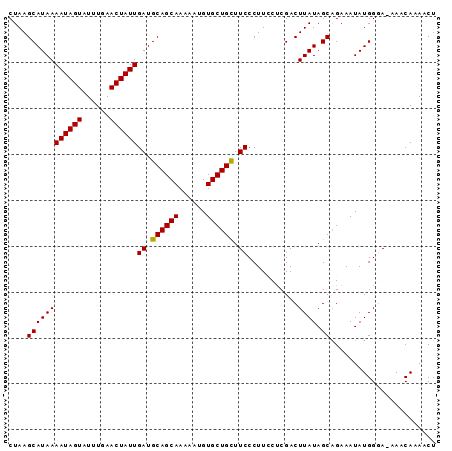
| Location | 15,809,201 – 15,809,293 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -18.60 |
| Consensus MFE | -15.68 |
| Energy contribution | -15.52 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.946919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15809201 92 + 22407834 CUAAGCAUAAAAUAGUAUUUGAACUAUUGAUGCAGCAAAAAUGUGCUGCUUCCCUUCCUCGACUUAUAGCAGAAAUAUGGGA-AAACAUAACU ..........((((((......))))))...((((((......))))))(((((....((..(.....)..)).....))))-)......... ( -18.70) >DroSec_CAF1 3751 92 + 1 CUAAGCAUAAAAUAGUAUUUGAACUAUUGAUGCAGCAAAAAUGUGCUGCUUCCCUUGCUCGACUUAUAGCAGAAAUAUGGGA-AAACAAAACU ..........((((((......))))))...((((((......))))))((((((((((........)))))......))))-)......... ( -20.40) >DroSim_CAF1 3765 92 + 1 CUAAGCAUAAAAUAGUAUUUGAACUAUUGAUGCAGCAAAAAUGUGCUGCUUCCCUUGCUCGACUUAUAGCAGAAAUAUGGGA-AAACAAAACU ..........((((((......))))))...((((((......))))))((((((((((........)))))......))))-)......... ( -20.40) >DroEre_CAF1 3841 93 + 1 CUAAGCAUAAAAUAGUAUUUGAACUAUUGAUGCAGCAAAAAUGUGCUGCUUCCCUUCCCCGACUUAUAGCAGAAAUAUGGCAAAAACAAAUCU ....((((((((((((......))))))((.((((((......)))))).))...........)))).))....................... ( -16.00) >DroYak_CAF1 28845 93 + 1 CUAAGCAUAAAAUAGUAUUUGAACUAUUGAUGCAGCAAAAAUGUGCUGUUUCCCUUCCCCGACUUAUAGCCGAAAUAUGGCAAAAACAAAACU .(((((....((((((......))))))((.((((((......)))))).))........).))))..((((.....))))............ ( -17.50) >consensus CUAAGCAUAAAAUAGUAUUUGAACUAUUGAUGCAGCAAAAAUGUGCUGCUUCCCUUCCUCGACUUAUAGCAGAAAUAUGGGA_AAACAAAACU ....((((((((((((......))))))((.((((((......)))))).))...........)))).))....................... (-15.68 = -15.52 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:51 2006