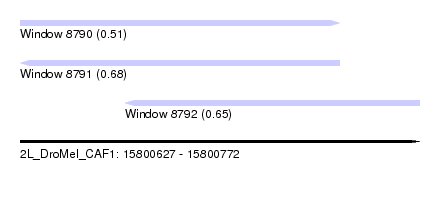
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,800,627 – 15,800,772 |
| Length | 145 |
| Max. P | 0.680541 |

| Location | 15,800,627 – 15,800,743 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 95.89 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -24.49 |
| Energy contribution | -24.80 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15800627 116 + 22407834 UCUGUUGCUAUUUCUUCCGCCGAAGAGGGUAACUUUUUGGGGCAUUGUCUGAUGACUGUCAGCUUACUGACGAUUUUUUUUU-CAGUUUUUAUUUUCAAUGAGUUGAGAUCGUGUUU- ......((.(((((..((.((((((((.....))))))))))(((((..(((.(((((((((....)))))((........)-))))).)))....)))))....))))).))....- ( -27.30) >DroSec_CAF1 24472 118 + 1 UCUGUUGCAAUUUCUUCCGCCGAAGAGGGUAACUUUUUGGGGCAUUGUCUGAUGACUGUCAGCUUACUGACGAUUUUUUUUUCCAGUUUUUAUUUUCAAUGAGUUGAGAUCGUGUUUU ......((.(((((..((.((((((((.....))))))))))(((((..(((.(((((((((....)))))((........)).)))).)))....)))))....))))).))..... ( -26.40) >DroSim_CAF1 15796 116 + 1 UCUGUUGCUAUUUCUUCCGCCGAAGAGGGUAACUUUUUGGGGCAUUGUCUGAUGACUGUCAGCUUACUGACGAUUUUUUUUU-CAGUUUUUAUUUUCAAUGAGUUGAGAUCGUGUUU- ......((.(((((..((.((((((((.....))))))))))(((((..(((.(((((((((....)))))((........)-))))).)))....)))))....))))).))....- ( -27.30) >DroEre_CAF1 24083 114 + 1 UCUGUUGCUAUUUCUUCCGCCGAAGAGGGUAACUUUUUGGGGCAUUGUCUGAUGACUGUCAGCUUACUGACGAUUUU--UUUCAGGUUUUUA--UUCAAUGAGUUGAGAUCGUGUUUU ......((.(((((..((.((((((((.....))))))))))(((((..(((.(((((((((....)))))((....--..)).)))).)))--..)))))....))))).))..... ( -29.70) >consensus UCUGUUGCUAUUUCUUCCGCCGAAGAGGGUAACUUUUUGGGGCAUUGUCUGAUGACUGUCAGCUUACUGACGAUUUUUUUUU_CAGUUUUUAUUUUCAAUGAGUUGAGAUCGUGUUU_ ......((.(((((..((.((((((((.....))))))))))(((((..(((.(((((((((....)))))((........)).)))).)))....)))))....))))).))..... (-24.49 = -24.80 + 0.31)

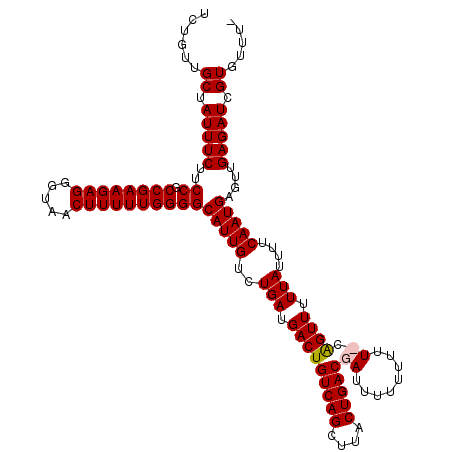
| Location | 15,800,627 – 15,800,743 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 95.89 |
| Mean single sequence MFE | -19.02 |
| Consensus MFE | -17.30 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.680541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15800627 116 - 22407834 -AAACACGAUCUCAACUCAUUGAAAAUAAAAACUG-AAAAAAAAAUCGUCAGUAAGCUGACAGUCAUCAGACAAUGCCCCAAAAAGUUACCCUCUUCGGCGGAAGAAAUAGCAACAGA -..........((((....)))).........(((-...........(((((....))))).(((....))).............((((...(((((....)))))..))))..))). ( -18.90) >DroSec_CAF1 24472 118 - 1 AAAACACGAUCUCAACUCAUUGAAAAUAAAAACUGGAAAAAAAAAUCGUCAGUAAGCUGACAGUCAUCAGACAAUGCCCCAAAAAGUUACCCUCUUCGGCGGAAGAAAUUGCAACAGA ......((((.((((((..(((.........................(((((....))))).(((....))).......)))..))))..((.(....).))..)).))))....... ( -19.60) >DroSim_CAF1 15796 116 - 1 -AAACACGAUCUCAACUCAUUGAAAAUAAAAACUG-AAAAAAAAAUCGUCAGUAAGCUGACAGUCAUCAGACAAUGCCCCAAAAAGUUACCCUCUUCGGCGGAAGAAAUAGCAACAGA -..........((((....)))).........(((-...........(((((....))))).(((....))).............((((...(((((....)))))..))))..))). ( -18.90) >DroEre_CAF1 24083 114 - 1 AAAACACGAUCUCAACUCAUUGAA--UAAAAACCUGAAA--AAAAUCGUCAGUAAGCUGACAGUCAUCAGACAAUGCCCCAAAAAGUUACCCUCUUCGGCGGAAGAAAUAGCAACAGA ...........((((....)))).--.......(((...--......(((((....))))).(((....))).............((((...(((((....)))))..))))..))). ( -18.70) >consensus _AAACACGAUCUCAACUCAUUGAAAAUAAAAACUG_AAAAAAAAAUCGUCAGUAAGCUGACAGUCAUCAGACAAUGCCCCAAAAAGUUACCCUCUUCGGCGGAAGAAAUAGCAACAGA .........(((.((((..(((.........................(((((....))))).(((....))).......)))..))))..((.(....).)).)))............ (-17.30 = -17.30 + 0.00)

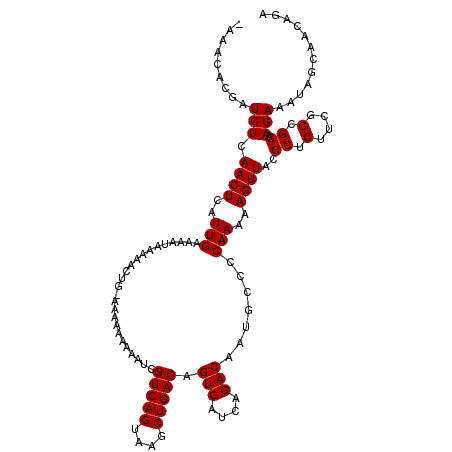
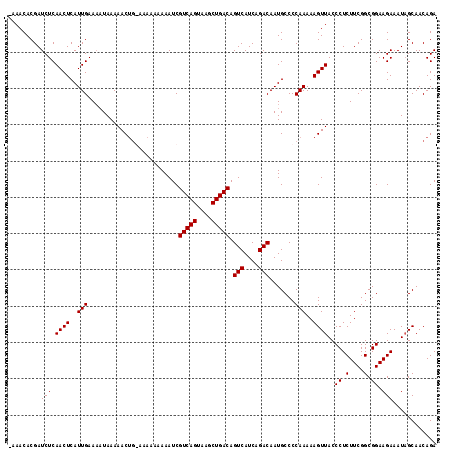
| Location | 15,800,665 – 15,800,772 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.29 |
| Mean single sequence MFE | -18.20 |
| Consensus MFE | -13.67 |
| Energy contribution | -14.23 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15800665 107 - 22407834 UCAAGGGUCGUUCGUCGGAUAUUAAAAA-----A------AAACACGAUCUCAACUCAUUGAAAAUAAAAACUG-AAAAAAAAAUCGUCAGUAAGCUGACAGUCAUCAGACAAUGCCCC ....(((.((((.(((.(((........-----.------..........((((....))))............-...........(((((....)))))....))).)))))))))). ( -20.50) >DroSim_CAF1 15834 112 - 1 UCAAAGGUCGUUCGUCGGAUAUUUAAAAAAAAAA------AAACACGAUCUCAACUCAUUGAAAAUAAAAACUG-AAAAAAAAAUCGUCAGUAAGCUGACAGUCAUCAGACAAUGCCCC .....((.((((.(((.(((..............------..........((((....))))............-...........(((((....)))))....))).))))))))).. ( -17.20) >DroEre_CAF1 24121 114 - 1 UCAGAGAUCCAUCGUCGGGUACUUAAAAAAACA-AAAACAAAACACGAUCUCAACUCAUUGAA--UAAAAACCUGAAA--AAAAUCGUCAGUAAGCUGACAGUCAUCAGACAAUGCCCC .....(((.....)))(((((............-.............................--.............--......(((((....))))).(((....)))..))))). ( -16.90) >consensus UCAAAGGUCGUUCGUCGGAUAUUUAAAAAAA_AA______AAACACGAUCUCAACUCAUUGAAAAUAAAAACUG_AAAAAAAAAUCGUCAGUAAGCUGACAGUCAUCAGACAAUGCCCC .....((.((((.(((.((((((...........................((((....))))........................(((((....)))))))).))).))))))).)). (-13.67 = -14.23 + 0.56)

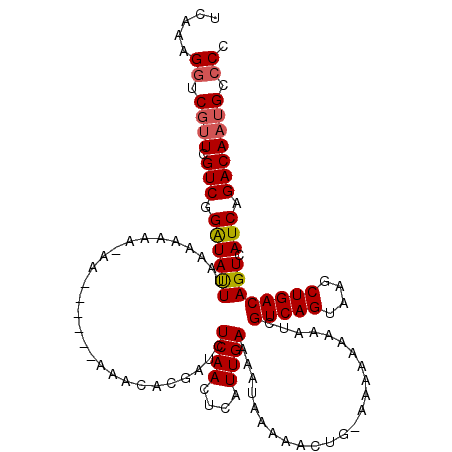
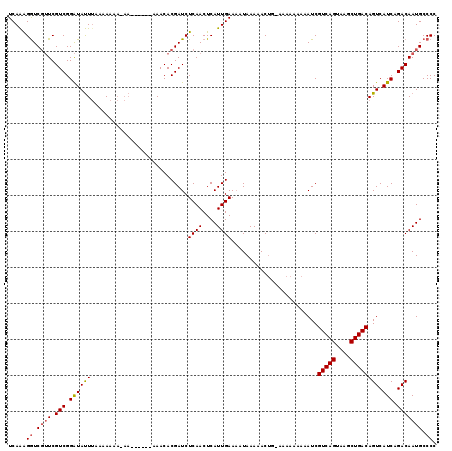
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:42 2006