
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,725,693 – 1,726,413 |
| Length | 720 |
| Max. P | 0.999129 |

| Location | 1,725,693 – 1,725,813 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -47.38 |
| Consensus MFE | -44.20 |
| Energy contribution | -44.16 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1725693 120 + 22407834 CGAUCUGGAUGUCGGUGCAGCCGGUCAGGUUGGACCAGGCCUCCCAGUGGUCCUGGUCAAAGGGAUCCUCAAUCGAGACGAUGGGGAAGUCCUUAAUAAACUCCUGGUACAGGUUGGCCA (((((((....((((.....))))(((((...(((((((((.......)).))))))).(((((.((((((.(((...)))))))))..)))))........)))))..))))))).... ( -46.90) >DroSec_CAF1 7846 120 + 1 CGAUCUGGAUAUCGGUGCAGCCGGUCAGGUUGGACCAGGCCUCCCAGUGGUCCUGGUCGAAGGGAUCCUCAAUCGAGACGAUGGGGAAGUCCUUGAUGAACUCCUGGUACAGGUCGGCCA (((((((...(((((.....)))))((((...(((((((((.......)).))))))).(((((.((((((.(((...)))))))))..)))))........))))...))))))).... ( -49.60) >DroSim_CAF1 8024 120 + 1 CGAUCUGGAUAUCGGUGCAGCCGGUCAGGUUGGACCAGGCCUCCCAGUGGUCCUGGUCAAAGGGAUCCUCAAUCGAGACGAUGGGGAAGUCCUUGAUGAACUCCUGGUACAGGUCGGCCA (((((((...(((((.....)))))((((...(((((((((.......)).))))))).(((((.((((((.(((...)))))))))..)))))........))))...))))))).... ( -49.50) >DroEre_CAF1 7872 120 + 1 CGAUCUGGAUCUGGGUGGAGCCGGUCAGGUUGCUCCAGGCCUCCCAGUGGUCCUGGUCGAAGGGAUCCUCGAUCGAGACGAUGGGGAAGUCCUUGAUGAACUCCUGGUACAGGUUGGCCA (((((((...(..((((((((..........))))))(((.((((.((.(((.(((((((.((...))))))))).))).)).)))).)))...........))..)..))))))).... ( -47.40) >DroYak_CAF1 8530 120 + 1 CGAUCUGGAUGCUGGUGGAGCCGGUCAGGUUGGUCCAGGCCUCCCAGUGGUCCUGGUCGAAGGGAUCCUCAAUCGAGACGAUGGGGAAGUCCUUGAUGAACUCUUGGUACAGGUUGGCCA (((((.(((((((((..(.(((((.(......).)).))))..))))).)))).)))))(((((.((((((.(((...)))))))))..)))))..........((((........)))) ( -43.50) >consensus CGAUCUGGAUAUCGGUGCAGCCGGUCAGGUUGGACCAGGCCUCCCAGUGGUCCUGGUCGAAGGGAUCCUCAAUCGAGACGAUGGGGAAGUCCUUGAUGAACUCCUGGUACAGGUUGGCCA (((((((....((((.....))))(((((...(((((((((.......)).))))))).(((((.((((((.(((...)))))))))..)))))........)))))..))))))).... (-44.20 = -44.16 + -0.04)

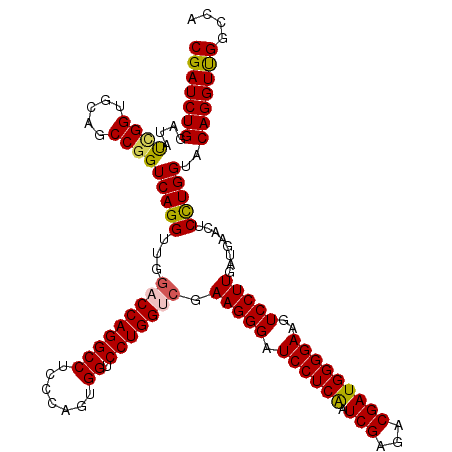
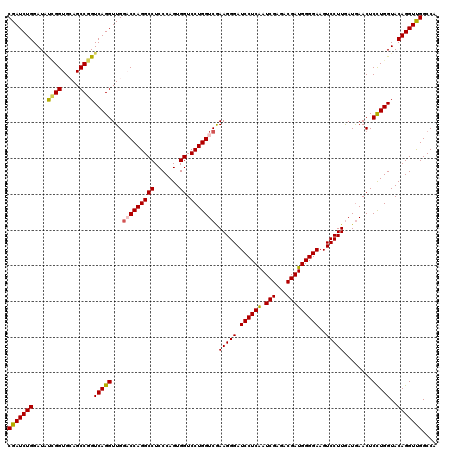
| Location | 1,725,773 – 1,725,893 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -40.42 |
| Consensus MFE | -36.38 |
| Energy contribution | -36.22 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1725773 120 - 22407834 GUUCUACAAGGAUGGCCAGUACGAUCUGGACUUCAAGAACGAGAAGAGCGACAAGUCGCAGUGGCUGCCCGCCGACAAGCUGGCCAACCUGUACCAGGAGUUUAUUAAGGACUUCCCCAU ....((((.((.((((((((.((.(((........))).))......((((....)))).(((((.....))).))..)))))))).))))))...(((((((.....)))))))..... ( -39.40) >DroSec_CAF1 7926 120 - 1 GUUCUACAGGGAUGGCCAGUACGAUCUGGACUUCAAGAACGAGAAGAGCGACAAGUCCCAGUGGCUGGCCGCCGACAAGCUGGCCGACCUGUACCAGGAGUUCAUCAAGGACUUCCCCAU ....((((((..((((((((.....((((.((((........))))...((....))))))((((.....))))....)))))))).))))))...(((((((.....)))))))..... ( -41.80) >DroSim_CAF1 8104 120 - 1 GUUCUACAGGGAUGGCCAGUACGAUCUGGACUUCAAGAACGAGAAGAGCGACAAGUCCCAGUGGCUGGCCGCCGACAAGCUGGCCGACCUGUACCAGGAGUUCAUCAAGGACUUCCCCAU ....((((((..((((((((.....((((.((((........))))...((....))))))((((.....))))....)))))))).))))))...(((((((.....)))))))..... ( -41.80) >DroEre_CAF1 7952 120 - 1 GUUCUACAAGGAUGGCCAGUACGAUCUGGACUUCAAGAACGAGAAGAGCGACAAGUCGCAGUGGCUGCCCGCCGACAAGCUGGCCAACCUGUACCAGGAGUUCAUCAAGGACUUCCCCAU ....((((.((.((((((((.((.(((........))).))......((((....)))).(((((.....))).))..)))))))).))))))...(((((((.....)))))))..... ( -41.40) >DroYak_CAF1 8610 120 - 1 GUUCUACAAGGAUGGCCAGUACGAUCUGGACUUCAAGAACGAGAAGAGCGACAAGGCGCAGUGGCUGCCCGCUGACAAGCUGGCCAACCUGUACCAAGAGUUCAUCAAGGACUUCCCCAU ....((((.((.((((((((.((.(((........))).)).....((((....(((((....)).))))))).....)))))))).))))))....((((((.....))))))...... ( -37.70) >consensus GUUCUACAAGGAUGGCCAGUACGAUCUGGACUUCAAGAACGAGAAGAGCGACAAGUCGCAGUGGCUGCCCGCCGACAAGCUGGCCAACCUGUACCAGGAGUUCAUCAAGGACUUCCCCAU ....((((.((.((((((((.....((((((((...(..(.....)..)...))))).)))((((.....))))....)))))))).))))))...(((((((.....)))))))..... (-36.38 = -36.22 + -0.16)

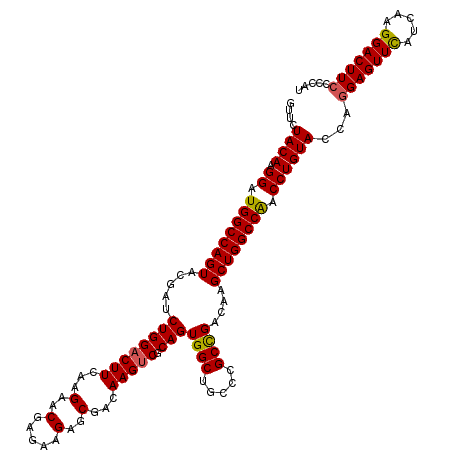
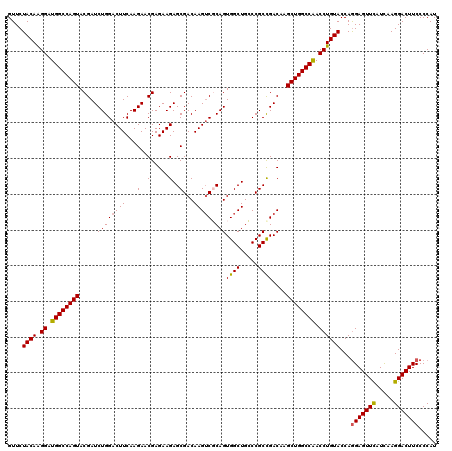
| Location | 1,725,853 – 1,725,973 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -44.62 |
| Consensus MFE | -43.54 |
| Energy contribution | -42.90 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1725853 120 - 22407834 CUGAACCUGAUCAGCGAUGCCAUCGCCAAGGCCGGAUACACCGGCAAGAUCGAGAUCGGCAUGGACGUGGCCGCCUCCGAGUUCUACAAGGAUGGCCAGUACGAUCUGGACUUCAAGAAC .((((((.((((.((.(((((((.(((...(((((.....)))))..(((....)))))))))).))).)).((((((..(.....)..))).)))......)))).))..))))..... ( -43.80) >DroSec_CAF1 8006 120 - 1 CUGAACCUGAUCAGCGAUGCCAUCGCCAAGGCCGGAUACACCGGCAAGAUUGAGAUCGGCAUGGACGUGGCCGCCUCUGAGUUCUACAGGGAUGGCCAGUACGAUCUGGACUUCAAGAAC .((((((.((((..(.((((((((..(((.(((((.....)))))....))).))).))))).)((.((((((.(((((.......))))).))))))))..)))).))..))))..... ( -47.70) >DroSim_CAF1 8184 120 - 1 CUGAACCUGAUCAGCGACGCCAUCGCCAAGGCCGGAUACACCGGCAAGAUUGAGAUCGGCAUGGACGUGGCCGCCUCCGAGUUCUACAGGGAUGGCCAGUACGAUCUGGACUUCAAGAAC .((((((.((((.((.(((((((.(((...(((((.....)))))..(((....)))))))))).))).)).((((((..(.....)..))).)))......)))).))..))))..... ( -44.20) >DroEre_CAF1 8032 120 - 1 CUGAACCUGAUCAGCGACGCCAUCGCCAAGGCCGGUUACACCGGCAAGAUCGAGAUCGGCAUGGACGUGGCCGCCUCUGAGUUCUACAAGGAUGGCCAGUACGAUCUGGACUUCAAGAAC .((((((.((((....(((((((.(((...(((((.....)))))..(((....)))))))))).)))((((((((.((.......))))).))))).....)))).))..))))..... ( -45.40) >DroYak_CAF1 8690 120 - 1 CUGAACCUGAUCAGCGAUGCCAUCGCCAAGGCCGGCUACACCGGCAAGAUCGAGAUCGGCAUGGAUGUGGCCGCCUCCGAGUUCUACAAGGAUGGCCAGUACGAUCUGGACUUCAAGAAC .((((((.((((.(((((...)))))...((((((((((((((((..(((....))).)).))).)))))))...(((..(.....)..))).)))).....)))).))..))))..... ( -42.00) >consensus CUGAACCUGAUCAGCGAUGCCAUCGCCAAGGCCGGAUACACCGGCAAGAUCGAGAUCGGCAUGGACGUGGCCGCCUCCGAGUUCUACAAGGAUGGCCAGUACGAUCUGGACUUCAAGAAC .((((((.((((.((.(((((((.(((...(((((.....)))))..(((....)))))))))).))).)).((((((..(.....)..))).)))......)))).))..))))..... (-43.54 = -42.90 + -0.64)

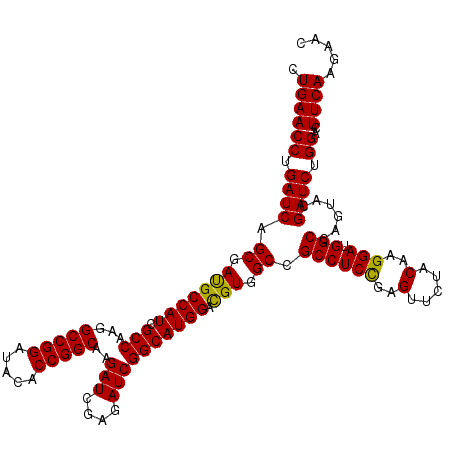
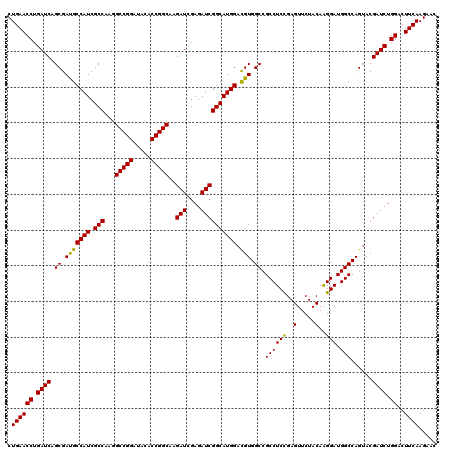
| Location | 1,725,893 – 1,726,013 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -48.68 |
| Consensus MFE | -48.72 |
| Energy contribution | -48.08 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.77 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1725893 120 - 22407834 GGGCGGCUUCGCCCCCAACAUCCAGUCCAACAAGGAGGCUCUGAACCUGAUCAGCGAUGCCAUCGCCAAGGCCGGAUACACCGGCAAGAUCGAGAUCGGCAUGGACGUGGCCGCCUCCGA (((((....)))))...................((((((..(((......)))((.(((((((.(((...(((((.....)))))..(((....)))))))))).))).)).)))))).. ( -49.20) >DroSec_CAF1 8046 120 - 1 GGGCGGCUUCGCCCCCAACAUCCAGUCCAACAAGGAGGCUCUGAACCUGAUCAGCGAUGCCAUCGCCAAGGCCGGAUACACCGGCAAGAUUGAGAUCGGCAUGGACGUGGCCGCCUCUGA (((((....)))))...................((((((..(((......)))((.(((((((.(((...(((((.....)))))..(((....)))))))))).))).)).)))))).. ( -45.90) >DroSim_CAF1 8224 120 - 1 GGGCGGCUUCGCCCCCAACAUCCAGUCCAACAAGGAGGCUCUGAACCUGAUCAGCGACGCCAUCGCCAAGGCCGGAUACACCGGCAAGAUUGAGAUCGGCAUGGACGUGGCCGCCUCCGA (((((....)))))...................((((((..(((......)))((.(((((((.(((...(((((.....)))))..(((....)))))))))).))).)).)))))).. ( -50.30) >DroEre_CAF1 8072 120 - 1 GGGCGGCUUCGCCCCCAACAUCCAGUCCAACAAGGAGGCCCUGAACCUGAUCAGCGACGCCAUCGCCAAGGCCGGUUACACCGGCAAGAUCGAGAUCGGCAUGGACGUGGCCGCCUCUGA (((((....)))))...................((((((..(((......)))((.(((((((.(((...(((((.....)))))..(((....)))))))))).))).)).)))))).. ( -48.80) >DroYak_CAF1 8730 120 - 1 GGGCGGCUUCGCCCCCAACAUCCAGUCCAACAAGGAGGCCCUGAACCUGAUCAGCGAUGCCAUCGCCAAGGCCGGCUACACCGGCAAGAUCGAGAUCGGCAUGGAUGUGGCCGCCUCCGA ((((((((.........(((((((.(((.....))).(((..((.(.(((((.(((((...)))))....(((((.....)))))..))))).).))))).))))))))))))))).... ( -49.20) >consensus GGGCGGCUUCGCCCCCAACAUCCAGUCCAACAAGGAGGCUCUGAACCUGAUCAGCGAUGCCAUCGCCAAGGCCGGAUACACCGGCAAGAUCGAGAUCGGCAUGGACGUGGCCGCCUCCGA (((((....)))))...................((((((..(((......)))((.(((((((.(((...(((((.....)))))..(((....)))))))))).))).)).)))))).. (-48.72 = -48.08 + -0.64)

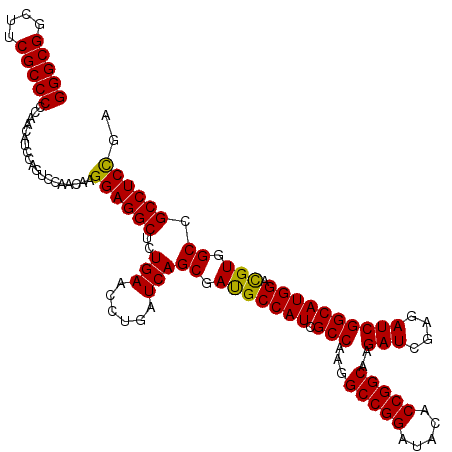
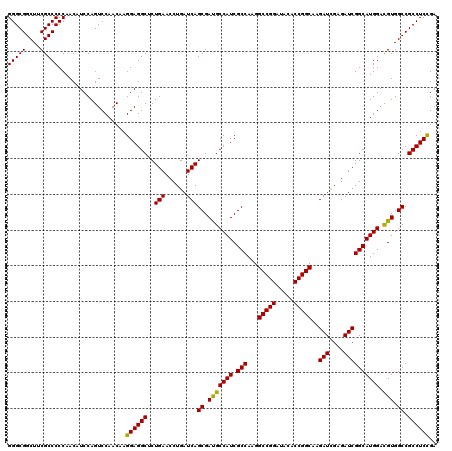
| Location | 1,725,933 – 1,726,053 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -56.62 |
| Consensus MFE | -56.00 |
| Energy contribution | -55.76 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.998154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1725933 120 + 22407834 UGUAUCCGGCCUUGGCGAUGGCAUCGCUGAUCAGGUUCAGAGCCUCCUUGUUGGACUGGAUGUUGGGGGCGAAGCCGCCCUCAUCGCCCACAGCGGUGGCGUCCAGACCGAACUUGGCCU .......(((((..(((((...)))))..)..((((((......(((.....)))((((((((((((((((....)))))))(((((.....))))))))))))))...)))))))))). ( -54.50) >DroSec_CAF1 8086 120 + 1 UGUAUCCGGCCUUGGCGAUGGCAUCGCUGAUCAGGUUCAGAGCCUCCUUGUUGGACUGGAUGUUGGGGGCGAAGCCGCCCUCAUCGCCCACGGCGGUGGCGUCCAGACCGAACUUGGCCU .......(((((..(((((...)))))..)..((((((......(((.....)))((((((((((((((((....)))))))((((((...)))))))))))))))...)))))))))). ( -56.70) >DroSim_CAF1 8264 120 + 1 UGUAUCCGGCCUUGGCGAUGGCGUCGCUGAUCAGGUUCAGAGCCUCCUUGUUGGACUGGAUGUUGGGGGCGAAGCCGCCCUCAUCGCCCACGGCGGUGGCGUCCAGACCGAACUUGGCCU .......(((((..(((((...)))))..)..((((((......(((.....)))((((((((((((((((....)))))))((((((...)))))))))))))))...)))))))))). ( -57.00) >DroEre_CAF1 8112 120 + 1 UGUAACCGGCCUUGGCGAUGGCGUCGCUGAUCAGGUUCAGGGCCUCCUUGUUGGACUGGAUGUUGGGGGCGAAGCCGCCCUCAUCGCCCACAGCGGUGGCGUCCAGACCGAACUUGGCCU .......(((((..(((((...)))))..)..((((((.((...(((.....)))((((((((((((((((....)))))))(((((.....)))))))))))))).)))))))))))). ( -56.50) >DroYak_CAF1 8770 120 + 1 UGUAGCCGGCCUUGGCGAUGGCAUCGCUGAUCAGGUUCAGGGCCUCCUUGUUGGACUGGAUGUUGGGGGCGAAGCCGCCCUCAUCGCCCACGGCGGUGGCGUCCAGACCGAACUUGGCCU .......(((((..(((((...)))))..)..((((((.((...(((.....)))((((((((((((((((....)))))))((((((...))))))))))))))).)))))))))))). ( -58.40) >consensus UGUAUCCGGCCUUGGCGAUGGCAUCGCUGAUCAGGUUCAGAGCCUCCUUGUUGGACUGGAUGUUGGGGGCGAAGCCGCCCUCAUCGCCCACGGCGGUGGCGUCCAGACCGAACUUGGCCU .......(((((..(((((...)))))..)..((((((......(((.....)))((((((((((((((((....)))))))(((((.....))))))))))))))...)))))))))). (-56.00 = -55.76 + -0.24)

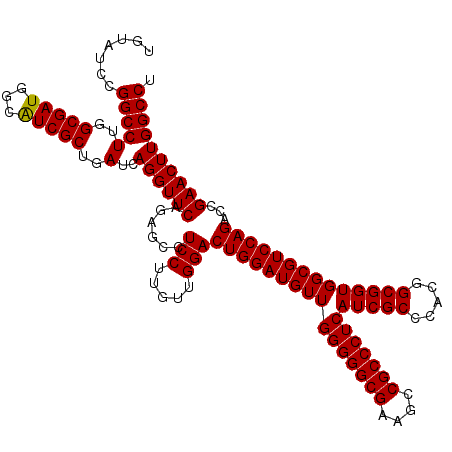
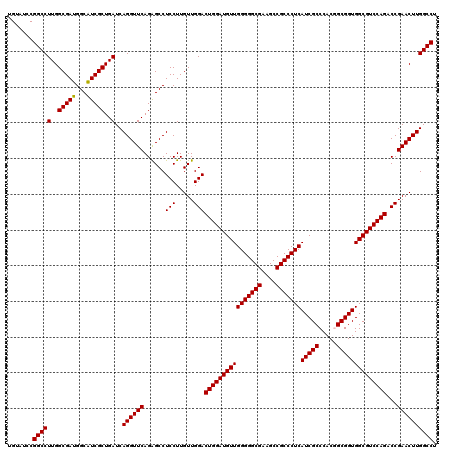
| Location | 1,725,933 – 1,726,053 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -48.06 |
| Consensus MFE | -47.14 |
| Energy contribution | -47.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1725933 120 - 22407834 AGGCCAAGUUCGGUCUGGACGCCACCGCUGUGGGCGAUGAGGGCGGCUUCGCCCCCAACAUCCAGUCCAACAAGGAGGCUCUGAACCUGAUCAGCGAUGCCAUCGCCAAGGCCGGAUACA .((((..(.(((((..(((.(((.((....(((((((((.(((((....)))))....))))..)))))....)).))))))..))).)).).(((((...)))))...))))....... ( -48.40) >DroSec_CAF1 8086 120 - 1 AGGCCAAGUUCGGUCUGGACGCCACCGCCGUGGGCGAUGAGGGCGGCUUCGCCCCCAACAUCCAGUCCAACAAGGAGGCUCUGAACCUGAUCAGCGAUGCCAUCGCCAAGGCCGGAUACA .((((..(.(((((..(((.(((.((....(((((((((.(((((....)))))....))))..)))))....)).))))))..))).)).).(((((...)))))...))))....... ( -48.40) >DroSim_CAF1 8264 120 - 1 AGGCCAAGUUCGGUCUGGACGCCACCGCCGUGGGCGAUGAGGGCGGCUUCGCCCCCAACAUCCAGUCCAACAAGGAGGCUCUGAACCUGAUCAGCGACGCCAUCGCCAAGGCCGGAUACA .((((..(.(((((..(((.(((.((....(((((((((.(((((....)))))....))))..)))))....)).))))))..))).)).).((((.....))))...))))....... ( -48.00) >DroEre_CAF1 8112 120 - 1 AGGCCAAGUUCGGUCUGGACGCCACCGCUGUGGGCGAUGAGGGCGGCUUCGCCCCCAACAUCCAGUCCAACAAGGAGGCCCUGAACCUGAUCAGCGACGCCAUCGCCAAGGCCGGUUACA .((((..(((.((.(((((((((((....)).)))).((.(((((....))))).))...))))).)))))..((.((((((((......)))).)..)))....))..))))....... ( -46.80) >DroYak_CAF1 8770 120 - 1 AGGCCAAGUUCGGUCUGGACGCCACCGCCGUGGGCGAUGAGGGCGGCUUCGCCCCCAACAUCCAGUCCAACAAGGAGGCCCUGAACCUGAUCAGCGAUGCCAUCGCCAAGGCCGGCUACA .((((..(((.((.(((((((((((....)).)))).((.(((((....))))).))...))))).))))).....((((.(((......)))(((((...)))))...))))))))... ( -48.70) >consensus AGGCCAAGUUCGGUCUGGACGCCACCGCCGUGGGCGAUGAGGGCGGCUUCGCCCCCAACAUCCAGUCCAACAAGGAGGCUCUGAACCUGAUCAGCGAUGCCAUCGCCAAGGCCGGAUACA .((((..(((.((.(((((((((((....)).)))).((.(((((....))))).))...))))).)))))..((.(((.((((......))))....)))....))..))))....... (-47.14 = -47.14 + 0.00)

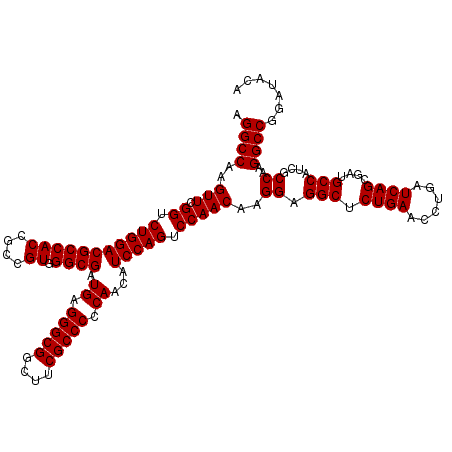
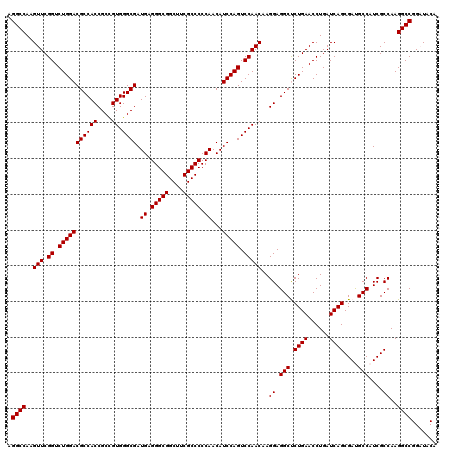
| Location | 1,725,973 – 1,726,093 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -54.60 |
| Consensus MFE | -52.24 |
| Energy contribution | -52.44 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1725973 120 + 22407834 AGCCUCCUUGUUGGACUGGAUGUUGGGGGCGAAGCCGCCCUCAUCGCCCACAGCGGUGGCGUCCAGACCGAACUUGGCCUUGAUCACAUUCUUCAGGUGGUGGUACACCUCGGAGCCCAU .(((......((((.((((((((((((((((....)))))))(((((.....)))))))))))))).))))....)))............(((((((((......))))).))))..... ( -50.60) >DroSec_CAF1 8126 120 + 1 AGCCUCCUUGUUGGACUGGAUGUUGGGGGCGAAGCCGCCCUCAUCGCCCACGGCGGUGGCGUCCAGACCGAACUUGGCCUUGAUCACGUUCUUCAGGUGGUGGUACACCUCGGAGCCCAU .(((......((((.((((((((((((((((....)))))))((((((...))))))))))))))).))))....))).........(((((..(((((......))))).))))).... ( -54.40) >DroSim_CAF1 8304 120 + 1 AGCCUCCUUGUUGGACUGGAUGUUGGGGGCGAAGCCGCCCUCAUCGCCCACGGCGGUGGCGUCCAGACCGAACUUGGCCUUGAUCACGUUCUUCAGGUGGUGGUACACCUCGGAGCCCAU .(((......((((.((((((((((((((((....)))))))((((((...))))))))))))))).))))....))).........(((((..(((((......))))).))))).... ( -54.40) >DroEre_CAF1 8152 120 + 1 GGCCUCCUUGUUGGACUGGAUGUUGGGGGCGAAGCCGCCCUCAUCGCCCACAGCGGUGGCGUCCAGACCGAACUUGGCCUUGAUCACGUUCUUCAGGUGGUGGUACACCUCGGAGCCCAU ((((......((((.((((((((((((((((....)))))))(((((.....)))))))))))))).))))....))))........(((((..(((((......))))).))))).... ( -55.70) >DroYak_CAF1 8810 120 + 1 GGCCUCCUUGUUGGACUGGAUGUUGGGGGCGAAGCCGCCCUCAUCGCCCACGGCGGUGGCGUCCAGACCGAACUUGGCCUUGAUCACGUUCUUCAGGUGGUGGUACACCUCGGAGCCCAU ((((......((((.((((((((((((((((....)))))))((((((...))))))))))))))).))))....))))........(((((..(((((......))))).))))).... ( -57.90) >consensus AGCCUCCUUGUUGGACUGGAUGUUGGGGGCGAAGCCGCCCUCAUCGCCCACGGCGGUGGCGUCCAGACCGAACUUGGCCUUGAUCACGUUCUUCAGGUGGUGGUACACCUCGGAGCCCAU .(((......((((.((((((((((((((((....)))))))(((((.....)))))))))))))).))))....))).........(((((..(((((......))))).))))).... (-52.24 = -52.44 + 0.20)

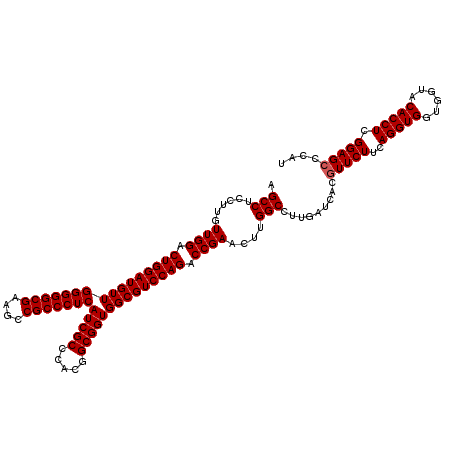
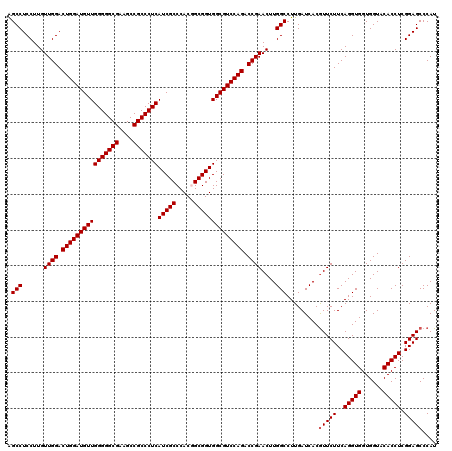
| Location | 1,725,973 – 1,726,093 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -47.58 |
| Consensus MFE | -47.98 |
| Energy contribution | -47.58 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.14 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1725973 120 - 22407834 AUGGGCUCCGAGGUGUACCACCACCUGAAGAAUGUGAUCAAGGCCAAGUUCGGUCUGGACGCCACCGCUGUGGGCGAUGAGGGCGGCUUCGCCCCCAACAUCCAGUCCAACAAGGAGGCU ..((.((((.(((((......)))))...((((.((........)).))))((.(((((((((((....)).)))).((.(((((....))))).))...))))).)).....)))).)) ( -45.80) >DroSec_CAF1 8126 120 - 1 AUGGGCUCCGAGGUGUACCACCACCUGAAGAACGUGAUCAAGGCCAAGUUCGGUCUGGACGCCACCGCCGUGGGCGAUGAGGGCGGCUUCGCCCCCAACAUCCAGUCCAACAAGGAGGCU ..((.((((.(((((......)))))...((((.((........)).))))((.(((((((((((....)).)))).((.(((((....))))).))...))))).)).....)))).)) ( -47.30) >DroSim_CAF1 8304 120 - 1 AUGGGCUCCGAGGUGUACCACCACCUGAAGAACGUGAUCAAGGCCAAGUUCGGUCUGGACGCCACCGCCGUGGGCGAUGAGGGCGGCUUCGCCCCCAACAUCCAGUCCAACAAGGAGGCU ..((.((((.(((((......)))))...((((.((........)).))))((.(((((((((((....)).)))).((.(((((....))))).))...))))).)).....)))).)) ( -47.30) >DroEre_CAF1 8152 120 - 1 AUGGGCUCCGAGGUGUACCACCACCUGAAGAACGUGAUCAAGGCCAAGUUCGGUCUGGACGCCACCGCUGUGGGCGAUGAGGGCGGCUUCGCCCCCAACAUCCAGUCCAACAAGGAGGCC ..((.((((.(((((......)))))...((((.((........)).))))((.(((((((((((....)).)))).((.(((((....))))).))...))))).)).....)))).)) ( -48.70) >DroYak_CAF1 8810 120 - 1 AUGGGCUCCGAGGUGUACCACCACCUGAAGAACGUGAUCAAGGCCAAGUUCGGUCUGGACGCCACCGCCGUGGGCGAUGAGGGCGGCUUCGCCCCCAACAUCCAGUCCAACAAGGAGGCC ..((.((((.(((((......)))))...((((.((........)).))))((.(((((((((((....)).)))).((.(((((....))))).))...))))).)).....)))).)) ( -48.80) >consensus AUGGGCUCCGAGGUGUACCACCACCUGAAGAACGUGAUCAAGGCCAAGUUCGGUCUGGACGCCACCGCCGUGGGCGAUGAGGGCGGCUUCGCCCCCAACAUCCAGUCCAACAAGGAGGCU ..((.((((.(((((......)))))...((((.((........)).))))((.(((((((((((....)).)))).((.(((((....))))).))...))))).)).....)))).)) (-47.98 = -47.58 + -0.40)

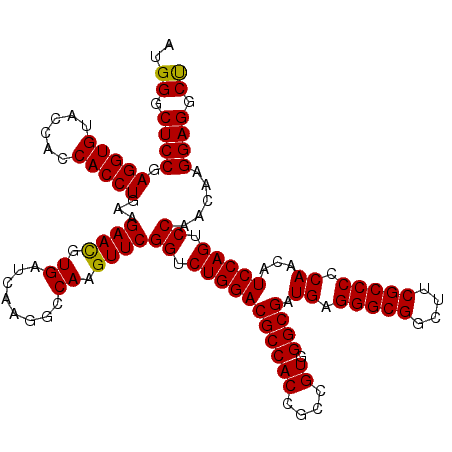
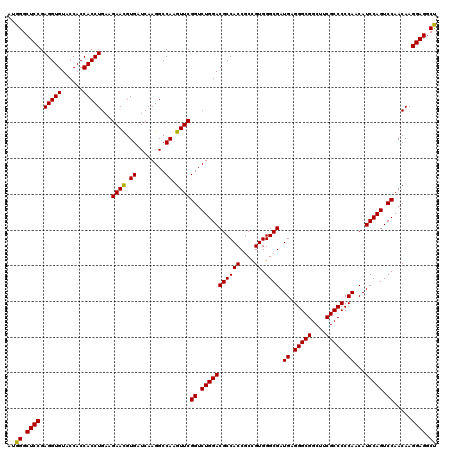
| Location | 1,726,013 – 1,726,133 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -53.59 |
| Consensus MFE | -53.14 |
| Energy contribution | -53.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1726013 120 + 22407834 UCAUCGCCCACAGCGGUGGCGUCCAGACCGAACUUGGCCUUGAUCACAUUCUUCAGGUGGUGGUACACCUCGGAGCCCAUCUUCAUGGCCUCUGUGAAGCUGGUGGCGCCGGUGGGCAGG .....((((((..(((((.((.((((.....((..((((.(((.......(((((((((......))))).)))).......))).))))...))....)))))).)))))))))))... ( -53.14) >DroSec_CAF1 8166 120 + 1 UCAUCGCCCACGGCGGUGGCGUCCAGACCGAACUUGGCCUUGAUCACGUUCUUCAGGUGGUGGUACACCUCGGAGCCCAUCUUCAUGGCCUCUGUGAAGCUGGUGGCGCCGGUGGGCAGG .....((((((..(((((.((.((((.....((..((((.(((....(..(((((((((......))))).))))..)....))).))))...))....)))))).)))))))))))... ( -53.70) >DroSim_CAF1 8344 120 + 1 UCAUCGCCCACGGCGGUGGCGUCCAGACCGAACUUGGCCUUGAUCACGUUCUUCAGGUGGUGGUACACCUCGGAGCCCAUCUUCAUGGCCUCUGUGAAGCUGGUGGCGCCGGUGGGCAGG .....((((((..(((((.((.((((.....((..((((.(((....(..(((((((((......))))).))))..)....))).))))...))....)))))).)))))))))))... ( -53.70) >DroEre_CAF1 8192 120 + 1 UCAUCGCCCACAGCGGUGGCGUCCAGACCGAACUUGGCCUUGAUCACGUUCUUCAGGUGGUGGUACACCUCGGAGCCCAUCUUCAUGGCCUCUGUGAAGCUGGUGGCGCCGGUGGGCAGG .....((((((..(((((.((.((((.....((..((((.(((....(..(((((((((......))))).))))..)....))).))))...))....)))))).)))))))))))... ( -53.70) >DroYak_CAF1 8850 120 + 1 UCAUCGCCCACGGCGGUGGCGUCCAGACCGAACUUGGCCUUGAUCACGUUCUUCAGGUGGUGGUACACCUCGGAGCCCAUCUUCAUGGCCUCUGUGAAGCUGGUGGCGCCGGUGGGCAGG .....((((((..(((((.((.((((.....((..((((.(((....(..(((((((((......))))).))))..)....))).))))...))....)))))).)))))))))))... ( -53.70) >consensus UCAUCGCCCACGGCGGUGGCGUCCAGACCGAACUUGGCCUUGAUCACGUUCUUCAGGUGGUGGUACACCUCGGAGCCCAUCUUCAUGGCCUCUGUGAAGCUGGUGGCGCCGGUGGGCAGG .....((((((..(((((.((.((((.....((..((((.(((.......(((((((((......))))).)))).......))).))))...))....)))))).)))))))))))... (-53.14 = -53.14 + 0.00)

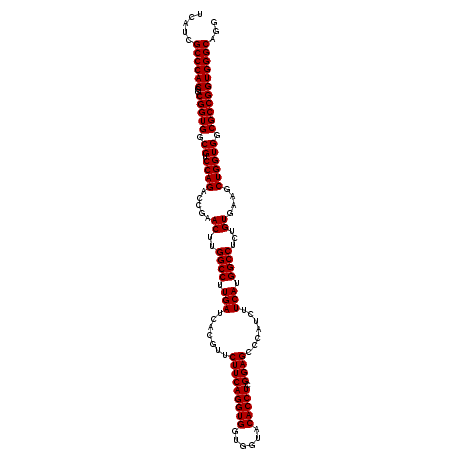
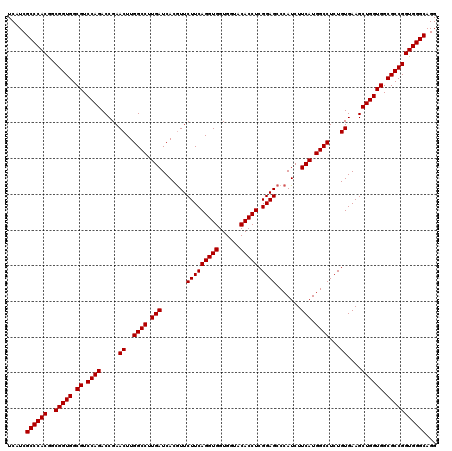
| Location | 1,726,013 – 1,726,133 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -48.72 |
| Consensus MFE | -48.96 |
| Energy contribution | -48.72 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.54 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1726013 120 - 22407834 CCUGCCCACCGGCGCCACCAGCUUCACAGAGGCCAUGAAGAUGGGCUCCGAGGUGUACCACCACCUGAAGAAUGUGAUCAAGGCCAAGUUCGGUCUGGACGCCACCGCUGUGGGCGAUGA ..(((((((.((((.(((...((((.(.(((.((((....)))).))).)(((((......)))))))))...))).((.(((((......))))).))......))))))))))).... ( -47.40) >DroSec_CAF1 8166 120 - 1 CCUGCCCACCGGCGCCACCAGCUUCACAGAGGCCAUGAAGAUGGGCUCCGAGGUGUACCACCACCUGAAGAACGUGAUCAAGGCCAAGUUCGGUCUGGACGCCACCGCCGUGGGCGAUGA ..(((((((.((((.(((...((((.(.(((.((((....)))).))).)(((((......)))))))))...))).((.(((((......))))).))......))))))))))).... ( -49.60) >DroSim_CAF1 8344 120 - 1 CCUGCCCACCGGCGCCACCAGCUUCACAGAGGCCAUGAAGAUGGGCUCCGAGGUGUACCACCACCUGAAGAACGUGAUCAAGGCCAAGUUCGGUCUGGACGCCACCGCCGUGGGCGAUGA ..(((((((.((((.(((...((((.(.(((.((((....)))).))).)(((((......)))))))))...))).((.(((((......))))).))......))))))))))).... ( -49.60) >DroEre_CAF1 8192 120 - 1 CCUGCCCACCGGCGCCACCAGCUUCACAGAGGCCAUGAAGAUGGGCUCCGAGGUGUACCACCACCUGAAGAACGUGAUCAAGGCCAAGUUCGGUCUGGACGCCACCGCUGUGGGCGAUGA ..(((((((.((((.(((...((((.(.(((.((((....)))).))).)(((((......)))))))))...))).((.(((((......))))).))......))))))))))).... ( -47.40) >DroYak_CAF1 8850 120 - 1 CCUGCCCACCGGCGCCACCAGCUUCACAGAGGCCAUGAAGAUGGGCUCCGAGGUGUACCACCACCUGAAGAACGUGAUCAAGGCCAAGUUCGGUCUGGACGCCACCGCCGUGGGCGAUGA ..(((((((.((((.(((...((((.(.(((.((((....)))).))).)(((((......)))))))))...))).((.(((((......))))).))......))))))))))).... ( -49.60) >consensus CCUGCCCACCGGCGCCACCAGCUUCACAGAGGCCAUGAAGAUGGGCUCCGAGGUGUACCACCACCUGAAGAACGUGAUCAAGGCCAAGUUCGGUCUGGACGCCACCGCCGUGGGCGAUGA ..(((((((.((((.(((...((((.(.(((.((((....)))).))).)(((((......)))))))))...))).((.(((((......))))).))......))))))))))).... (-48.96 = -48.72 + -0.24)

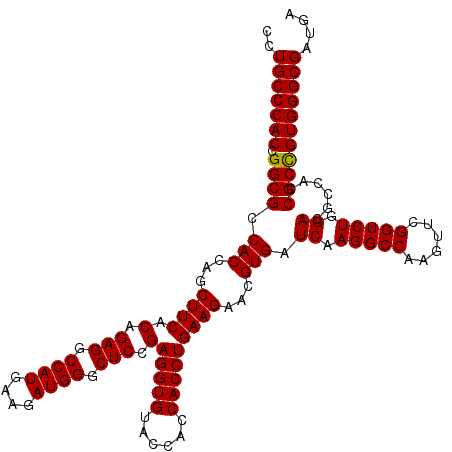
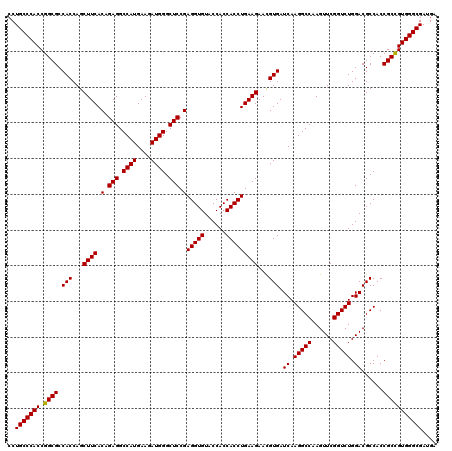
| Location | 1,726,053 – 1,726,173 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -50.60 |
| Consensus MFE | -50.32 |
| Energy contribution | -50.36 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1726053 120 + 22407834 UGAUCACAUUCUUCAGGUGGUGGUACACCUCGGAGCCCAUCUUCAUGGCCUCUGUGAAGCUGGUGGCGCCGGUGGGCAGGAUCAUGAACUCCUGCAUGGCCAGCUUGUUGCCGGCGUGGC ...((((....((((((((......)))).(((((.((((....)))).)))))))))(((((..((((.(((.(((((((........)))))).).))).))..))..))))))))). ( -51.70) >DroSec_CAF1 8206 120 + 1 UGAUCACGUUCUUCAGGUGGUGGUACACCUCGGAGCCCAUCUUCAUGGCCUCUGUGAAGCUGGUGGCGCCGGUGGGCAGGAUCAUGAACUCCUGCAUGGCCAGCUUGUUGCCGGCGUGGC ...((((....((((((((......)))).(((((.((((....)))).)))))))))(((((..((((.(((.(((((((........)))))).).))).))..))..))))))))). ( -51.70) >DroSim_CAF1 8384 120 + 1 UGAUCACGUUCUUCAGGUGGUGGUACACCUCGGAGCCCAUCUUCAUGGCCUCUGUGAAGCUGGUGGCGCCGGUGGGCAGGAUCAUGAACUCCUGCAUGGCAAGCUUGUUGCCGGCGUGGC ...((((....((((((((......)))).(((((.((((....)))).)))))))))(((((..((((((....((((((........)))))).))))......))..))))))))). ( -48.90) >DroEre_CAF1 8232 120 + 1 UGAUCACGUUCUUCAGGUGGUGGUACACCUCGGAGCCCAUCUUCAUGGCCUCUGUGAAGCUGGUGGCGCCGGUGGGCAGGAUCAUGAACUCCUGCAUGGCCAGCUUGUUGCCGGCGUGGC ...((((....((((((((......)))).(((((.((((....)))).)))))))))(((((..((((.(((.(((((((........)))))).).))).))..))..))))))))). ( -51.70) >DroYak_CAF1 8890 120 + 1 UGAUCACGUUCUUCAGGUGGUGGUACACCUCGGAGCCCAUCUUCAUGGCCUCUGUGAAGCUGGUGGCGCCGGUGGGCAGGAUCAUGAACUCUUGCAUGGCCAGCUUGUUGCCGGCGUGGC ...((((....((((((((......)))).(((((.((((....)))).)))))))))(((((..((((.(((.(((((((........)))))).).))).))..))..))))))))). ( -49.00) >consensus UGAUCACGUUCUUCAGGUGGUGGUACACCUCGGAGCCCAUCUUCAUGGCCUCUGUGAAGCUGGUGGCGCCGGUGGGCAGGAUCAUGAACUCCUGCAUGGCCAGCUUGUUGCCGGCGUGGC ...((((....((((((((......)))).(((((.((((....)))).)))))))))(((((..((((.(((.(((((((........)))))).).))).))..))..))))))))). (-50.32 = -50.36 + 0.04)

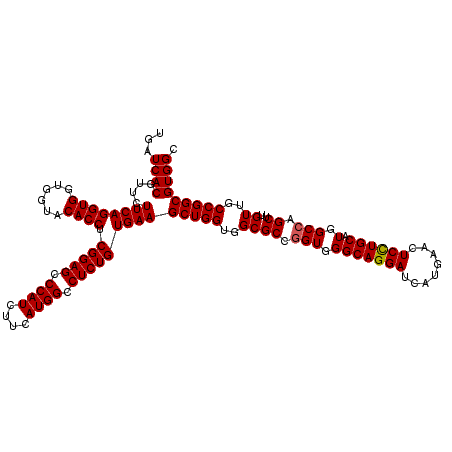
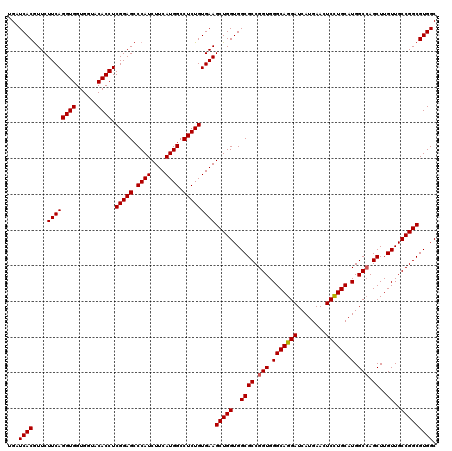
| Location | 1,726,053 – 1,726,173 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -46.76 |
| Consensus MFE | -46.18 |
| Energy contribution | -46.58 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.999129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1726053 120 - 22407834 GCCACGCCGGCAACAAGCUGGCCAUGCAGGAGUUCAUGAUCCUGCCCACCGGCGCCACCAGCUUCACAGAGGCCAUGAAGAUGGGCUCCGAGGUGUACCACCACCUGAAGAAUGUGAUCA (((..((((((.....))))))...((((((........)))))).....)))..(((...((((.(.(((.((((....)))).))).)(((((......)))))))))...))).... ( -48.60) >DroSec_CAF1 8206 120 - 1 GCCACGCCGGCAACAAGCUGGCCAUGCAGGAGUUCAUGAUCCUGCCCACCGGCGCCACCAGCUUCACAGAGGCCAUGAAGAUGGGCUCCGAGGUGUACCACCACCUGAAGAACGUGAUCA (((..((((((.....))))))...((((((........)))))).....)))..(((...((((.(.(((.((((....)))).))).)(((((......)))))))))...))).... ( -48.60) >DroSim_CAF1 8384 120 - 1 GCCACGCCGGCAACAAGCUUGCCAUGCAGGAGUUCAUGAUCCUGCCCACCGGCGCCACCAGCUUCACAGAGGCCAUGAAGAUGGGCUCCGAGGUGUACCACCACCUGAAGAACGUGAUCA ....(((((((((.....)))))..((((((........)))))).....)))).(((...((((.(.(((.((((....)))).))).)(((((......)))))))))...))).... ( -45.90) >DroEre_CAF1 8232 120 - 1 GCCACGCCGGCAACAAGCUGGCCAUGCAGGAGUUCAUGAUCCUGCCCACCGGCGCCACCAGCUUCACAGAGGCCAUGAAGAUGGGCUCCGAGGUGUACCACCACCUGAAGAACGUGAUCA (((..((((((.....))))))...((((((........)))))).....)))..(((...((((.(.(((.((((....)))).))).)(((((......)))))))))...))).... ( -48.60) >DroYak_CAF1 8890 120 - 1 GCCACGCCGGCAACAAGCUGGCCAUGCAAGAGUUCAUGAUCCUGCCCACCGGCGCCACCAGCUUCACAGAGGCCAUGAAGAUGGGCUCCGAGGUGUACCACCACCUGAAGAACGUGAUCA (((..((((((.....))))))...(((.((........)).))).....)))..(((...((((.(.(((.((((....)))).))).)(((((......)))))))))...))).... ( -42.10) >consensus GCCACGCCGGCAACAAGCUGGCCAUGCAGGAGUUCAUGAUCCUGCCCACCGGCGCCACCAGCUUCACAGAGGCCAUGAAGAUGGGCUCCGAGGUGUACCACCACCUGAAGAACGUGAUCA (((..((((((.....))))))...((((((........)))))).....)))..(((...((((.(.(((.((((....)))).))).)(((((......)))))))))...))).... (-46.18 = -46.58 + 0.40)

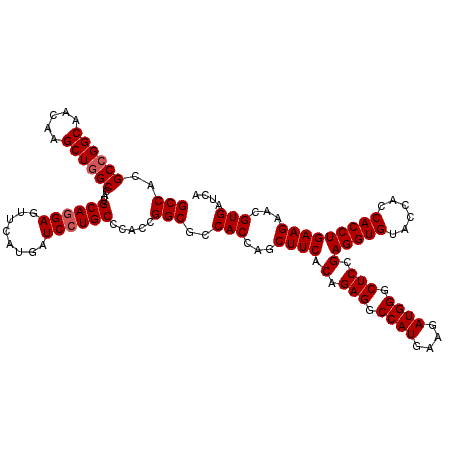
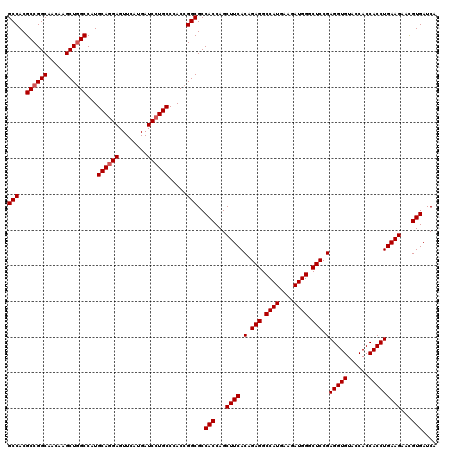
| Location | 1,726,093 – 1,726,213 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.42 |
| Mean single sequence MFE | -44.98 |
| Consensus MFE | -41.56 |
| Energy contribution | -42.16 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1726093 120 - 22407834 AUCCUGCCGGUGCCCGCUUUCAACGUGAUCAACGGCGGCAGCCACGCCGGCAACAAGCUGGCCAUGCAGGAGUUCAUGAUCCUGCCCACCGGCGCCACCAGCUUCACAGAGGCCAUGAAG .....((((.((..(((.......)))..)).))))(((.(((..((((((.....))))))...((((((........)))))).....))))))....(((((...)))))....... ( -45.60) >DroSec_CAF1 8246 120 - 1 AUCCUGCCGGUUCCCGCUUUCAACGUGAUCAACGGCGGCAGCCACGCCGGCAACAAGCUGGCCAUGCAGGAGUUCAUGAUCCUGCCCACCGGCGCCACCAGCUUCACAGAGGCCAUGAAG .((((((.((((.(((((.((.....)).....))))).))))..((((((.....))))))...)))))).((((((.....(((....))).......(((((...))))))))))). ( -46.50) >DroSim_CAF1 8424 120 - 1 AUCCUGCCGGUGCCCGCUUUCAACGUGAUCAACGGCGGCAGCCACGCCGGCAACAAGCUUGCCAUGCAGGAGUUCAUGAUCCUGCCCACCGGCGCCACCAGCUUCACAGAGGCCAUGAAG .((((((.((((...(((.((..(((.....)))..)).)))..))))(((((.....)))))..)))))).((((((.....(((....))).......(((((...))))))))))). ( -42.30) >DroEre_CAF1 8272 120 - 1 AUCCUGCCGGUGCCCGCUUUCAACGUGAUCAACGGUGGCAGCCACGCCGGCAACAAGCUGGCCAUGCAGGAGUUCAUGAUCCUGCCCACCGGCGCCACCAGCUUCACAGAGGCCAUGAAG .....(((.(((...(((......((.....))((((((.(((..((((((.....))))))...((((((........)))))).....))))))))))))..)))...)))....... ( -49.00) >DroYak_CAF1 8930 120 - 1 AUCCUGCCGGUGCCCGCUUUCAACGUGAUCAACGGAGGCAGCCACGCCGGCAACAAGCUGGCCAUGCAAGAGUUCAUGAUCCUGCCCACCGGCGCCACCAGCUUCACAGAGGCCAUGAAG .....(((((((.(((...((.....))....))).(((((....((((((.....))))))((((........))))...)))))))))))).......(((((...)))))....... ( -41.50) >consensus AUCCUGCCGGUGCCCGCUUUCAACGUGAUCAACGGCGGCAGCCACGCCGGCAACAAGCUGGCCAUGCAGGAGUUCAUGAUCCUGCCCACCGGCGCCACCAGCUUCACAGAGGCCAUGAAG .....(((.(((...(((......((.....))((.(((.(((..((((((.....))))))...((((((........)))))).....)))))).)))))..)))...)))....... (-41.56 = -42.16 + 0.60)

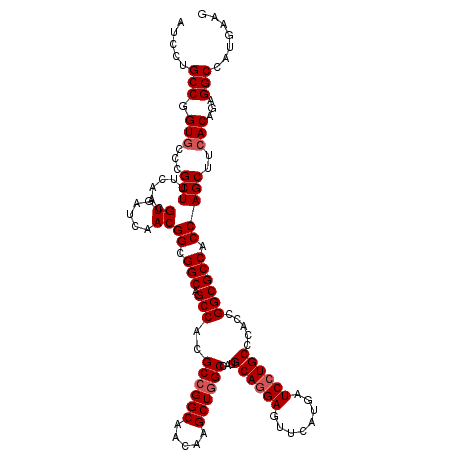
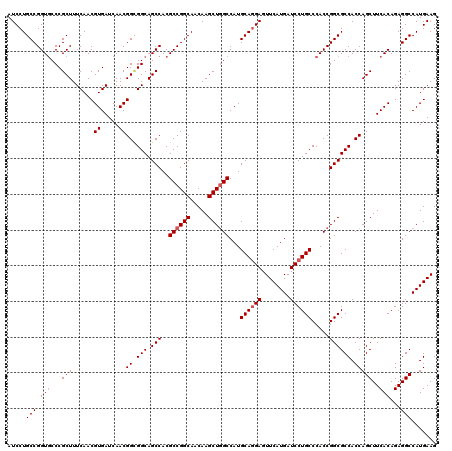
| Location | 1,726,133 – 1,726,253 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.75 |
| Mean single sequence MFE | -41.26 |
| Consensus MFE | -38.88 |
| Energy contribution | -39.12 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1726133 120 - 22407834 GUACAAACACAUUGCCGAUCUGGCUGGCAACAAGGAGAUCAUCCUGCCGGUGCCCGCUUUCAACGUGAUCAACGGCGGCAGCCACGCCGGCAACAAGCUGGCCAUGCAGGAGUUCAUGAU .........(((..((....((((..((....((((.....))))(((((((...(((.((..(((.....)))..)).)))..))))))).....))..))))....)).....))).. ( -41.70) >DroSec_CAF1 8286 120 - 1 GUACAAACACAUUGCCGAUCUGGCUGGCAACAAGGAGAUCAUCCUGCCGGUUCCCGCUUUCAACGUGAUCAACGGCGGCAGCCACGCCGGCAACAAGCUGGCCAUGCAGGAGUUCAUGAU .........(((.((.(((((...((....))...))))).((((((.((((.(((((.((.....)).....))))).))))..((((((.....))))))...))))))))..))).. ( -41.60) >DroSim_CAF1 8464 120 - 1 GUACAAACACAUCGCCGAUCUGGCUGGCAACAAGGAGAUCAUCCUGCCGGUGCCCGCUUUCAACGUGAUCAACGGCGGCAGCCACGCCGGCAACAAGCUUGCCAUGCAGGAGUUCAUGAU ..........((((....(((.(((((((((.((((.....))))(((((((...(((.((..(((.....)))..)).)))..))))))).....).)))))).)).))).....)))) ( -39.20) >DroEre_CAF1 8312 120 - 1 GUACAAACACAUUGCCGAUCUGGCUGGCAACAAGGAGAUCAUCCUGCCGGUGCCCGCUUUCAACGUGAUCAACGGUGGCAGCCACGCCGGCAACAAGCUGGCCAUGCAGGAGUUCAUGAU .........(((..((....((((..((....((((.....))))(((((((...(((.(((.(((.....))).))).)))..))))))).....))..))))....)).....))).. ( -42.80) >DroYak_CAF1 8970 120 - 1 GUACAAACACAUUGCCGAUCUGGCUGGCAACAAGGACAUCAUCCUGCCGGUGCCCGCUUUCAACGUGAUCAACGGAGGCAGCCACGCCGGCAACAAGCUGGCCAUGCAAGAGUUCAUGAU .....(((...((((.....((((..((....((((.....))))(((((((...(((.((..(((.....)))..)).)))..))))))).....))..)))).))))..)))...... ( -41.00) >consensus GUACAAACACAUUGCCGAUCUGGCUGGCAACAAGGAGAUCAUCCUGCCGGUGCCCGCUUUCAACGUGAUCAACGGCGGCAGCCACGCCGGCAACAAGCUGGCCAUGCAGGAGUUCAUGAU .....(((...((((.....((((..((....((((.....))))(((((((...(((.((..(((.....)))..)).)))..))))))).....))..)))).))))..)))...... (-38.88 = -39.12 + 0.24)

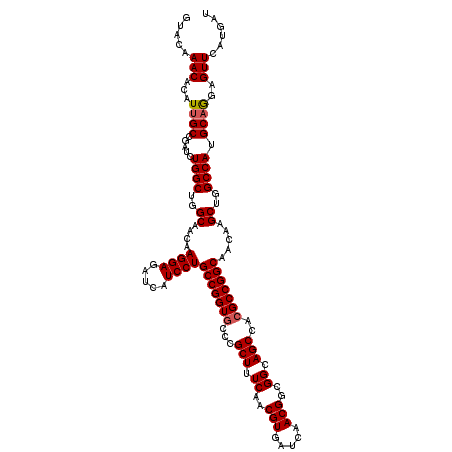
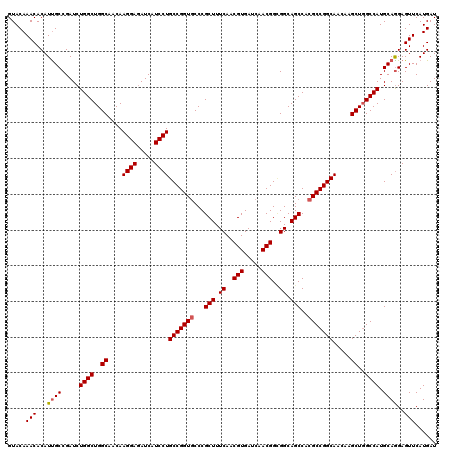
| Location | 1,726,253 – 1,726,373 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -51.76 |
| Consensus MFE | -48.20 |
| Energy contribution | -48.60 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1726253 120 + 22407834 AGCGGCACGCCCUUCUUGGCGGCUCCAGCCUUGGCCACGGCCAGGGAAACGCCCAGGAUGGCGUUGGCUCCGAACUUGCUCUUGUUCUCGGUGCCGUCCAACUUGAUCAUGAAGUUGUCA ...((((((((.((((.((((.......(((((((....)))))))...)))).)))).))))).(((.((((....((....))..)))).))))))((((((.......))))))... ( -51.20) >DroSec_CAF1 8406 120 + 1 AGGGGCACGCCCUUCUUGGCGGCUCCGGCCUUGGCCACGGCCAGGGAAACGCCCAGGAUGGCGUUGGCUCCGAACUUGCUCUUGUUCUCUGUGCCGUCCAACUUGAUCAUGAAGUUGUCG .(((((.((((......)))))))))(((....)))(((((((((((((((((......))))))...)))((((........)))).))).))))).((((((.......))))))... ( -52.00) >DroSim_CAF1 8584 120 + 1 AGGGGCACGCCCUUCUUGGCGGCUCCGGCCUUGGCCACGGCCAGGGAAACGCCCAGGAUGGCGUUGGCUCCGAACUUGCUCUUGUUCUCGGUGCCGUCCAACUUGAUCAUGAAGUUGUCG .(((((.((((......)))))))))..(((((((....))))))).((((((......))))))(((.((((....((....))..)))).)))...((((((.......))))))... ( -53.60) >DroEre_CAF1 8432 120 + 1 AGCGGCACGGCCUUCUUGGCGGCACCAGCCUUGGCCACGGCCAGGGAAACGCCCAGGAUGGCGUUGGCUCCGAACUUGCUCUUGUUCUCGGUGCCGUCCAACUUGAUCAUGAAGUUGUCG .(((((((.(((.....)))((..((..(((((((....))))))).((((((......))))))))..))((((........))))...))))))).((((((.......))))))... ( -50.80) >DroYak_CAF1 9090 120 + 1 AGCGGCACGCCCUUCUUGGCGGCACCAGCCUUGGCCACGGCCAGGGAAACGCCCAGGAUGGCGUUGGCUCCGAACUUGCUCUUGUUCUCGGUGCCGUCCAACUUGAUCAUGAAGUUGUCG ...((((((((.((((.((((.......(((((((....)))))))...)))).)))).))))).(((.((((....((....))..)))).))))))((((((.......))))))... ( -51.20) >consensus AGCGGCACGCCCUUCUUGGCGGCUCCAGCCUUGGCCACGGCCAGGGAAACGCCCAGGAUGGCGUUGGCUCCGAACUUGCUCUUGUUCUCGGUGCCGUCCAACUUGAUCAUGAAGUUGUCG ...((((((((.((((.((((.......(((((((....)))))))...)))).)))).))))).(((.((((....((....))..)))).))))))((((((.......))))))... (-48.20 = -48.60 + 0.40)

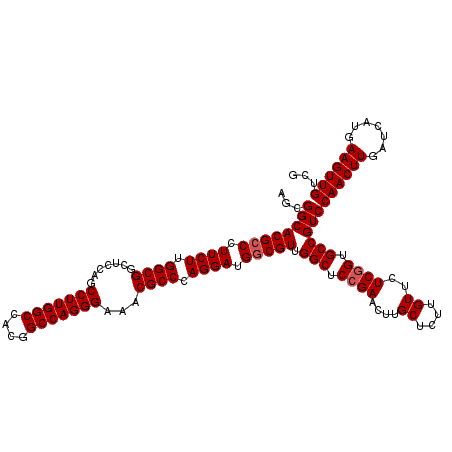
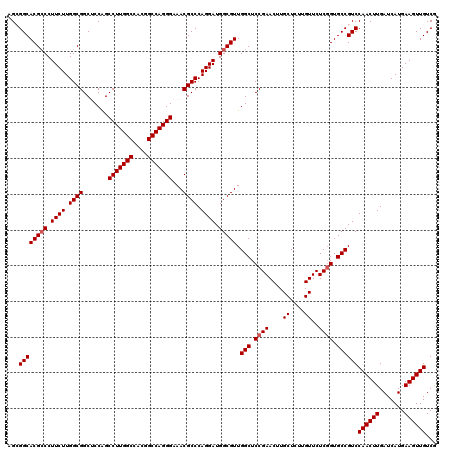
| Location | 1,726,253 – 1,726,373 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -47.56 |
| Consensus MFE | -43.60 |
| Energy contribution | -43.80 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1726253 120 - 22407834 UGACAACUUCAUGAUCAAGUUGGACGGCACCGAGAACAAGAGCAAGUUCGGAGCCAACGCCAUCCUGGGCGUUUCCCUGGCCGUGGCCAAGGCUGGAGCCGCCAAGAAGGGCGUGCCGCU ...((((((.......))))))..((((((...((((........))))((..((((((((......)))))).((.((((....)))).))..))..))(((......))))))))).. ( -46.50) >DroSec_CAF1 8406 120 - 1 CGACAACUUCAUGAUCAAGUUGGACGGCACAGAGAACAAGAGCAAGUUCGGAGCCAACGCCAUCCUGGGCGUUUCCCUGGCCGUGGCCAAGGCCGGAGCCGCCAAGAAGGGCGUGCCCCU ...((((((.......)))))).(((((.(((.((((........))))(((...((((((......)))))))))))))))))(((....)))((.((((((......)))).)).)). ( -48.90) >DroSim_CAF1 8584 120 - 1 CGACAACUUCAUGAUCAAGUUGGACGGCACCGAGAACAAGAGCAAGUUCGGAGCCAACGCCAUCCUGGGCGUUUCCCUGGCCGUGGCCAAGGCCGGAGCCGCCAAGAAGGGCGUGCCCCU ...((((((.......)))))).(((((.....((((........))))(((...((((((......)))))))))...)))))(((....)))((.((((((......)))).)).)). ( -45.50) >DroEre_CAF1 8432 120 - 1 CGACAACUUCAUGAUCAAGUUGGACGGCACCGAGAACAAGAGCAAGUUCGGAGCCAACGCCAUCCUGGGCGUUUCCCUGGCCGUGGCCAAGGCUGGUGCCGCCAAGAAGGCCGUGCCGCU .........((((..(...((((.(((((((..((((........))))..((((((((((......))))))....((((....)))).)))))))))))))))...)..))))..... ( -48.60) >DroYak_CAF1 9090 120 - 1 CGACAACUUCAUGAUCAAGUUGGACGGCACCGAGAACAAGAGCAAGUUCGGAGCCAACGCCAUCCUGGGCGUUUCCCUGGCCGUGGCCAAGGCUGGUGCCGCCAAGAAGGGCGUGCCGCU ......((((.........((((.(((((((..((((........))))..((((((((((......))))))....((((....)))).)))))))))))))))))))((((...)))) ( -48.30) >consensus CGACAACUUCAUGAUCAAGUUGGACGGCACCGAGAACAAGAGCAAGUUCGGAGCCAACGCCAUCCUGGGCGUUUCCCUGGCCGUGGCCAAGGCUGGAGCCGCCAAGAAGGGCGUGCCGCU ...((((((.......))))))...(((((...((((........))))((..((((((((......)))))).((.((((....)))).))..))..))(((......))))))))... (-43.60 = -43.80 + 0.20)

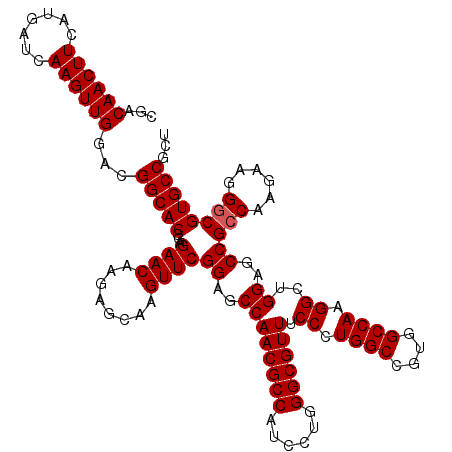
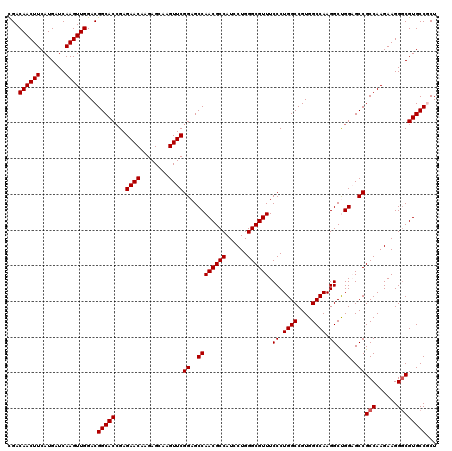
| Location | 1,726,293 – 1,726,413 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -41.84 |
| Consensus MFE | -40.12 |
| Energy contribution | -40.48 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1726293 120 + 22407834 CCAGGGAAACGCCCAGGAUGGCGUUGGCUCCGAACUUGCUCUUGUUCUCGGUGCCGUCCAACUUGAUCAUGAAGUUGUCAAUGGACGCCUGGUCAACCACAUCCAGAUUGGCCUUGAUCA ...(((.....))).(((((..(((((((..((((........))))..((((((((.((((((.......))))))...)))).)))).)))))))..))))).((((......)))). ( -40.20) >DroSec_CAF1 8446 120 + 1 CCAGGGAAACGCCCAGGAUGGCGUUGGCUCCGAACUUGCUCUUGUUCUCUGUGCCGUCCAACUUGAUCAUGAAGUUGUCGAUGGACGCUUGGUCAACCACAUCCAGAUUGGCCUUGAUCA ...(((.....))).(((((..(((((((..((((........))))...((((((((((((((.......))))))..))))).)))..)))))))..))))).((((......)))). ( -39.60) >DroSim_CAF1 8624 120 + 1 CCAGGGAAACGCCCAGGAUGGCGUUGGCUCCGAACUUGCUCUUGUUCUCGGUGCCGUCCAACUUGAUCAUGAAGUUGUCGAUGGACGCCUGGUCAACCACAUCCAGAUUGGCCUUGAUCA ...(((.....))).(((((..(((((((..((((........))))..(((((((((((((((.......))))))..))))).)))).)))))))..))))).((((......)))). ( -43.20) >DroEre_CAF1 8472 120 + 1 CCAGGGAAACGCCCAGGAUGGCGUUGGCUCCGAACUUGCUCUUGUUCUCGGUGCCGUCCAACUUGAUCAUGAAGUUGUCGAUGGACGCCUGGUCGACCACAUCCAGAUUGGCCUUGAUCA ...(((.....))).(((((..(((((((..((((........))))..(((((((((((((((.......))))))..))))).)))).)))))))..))))).((((......)))). ( -42.90) >DroYak_CAF1 9130 120 + 1 CCAGGGAAACGCCCAGGAUGGCGUUGGCUCCGAACUUGCUCUUGUUCUCGGUGCCGUCCAACUUGAUCAUGAAGUUGUCGAUGGCAGCCUGGUCGACCACAUCCAGAUUGGCCUUGAUCA ...(((.....))).(((((..(((((((..((((........))))..(((((((((((((((.......))))))..))))))).)).)))))))..))))).((((......)))). ( -43.30) >consensus CCAGGGAAACGCCCAGGAUGGCGUUGGCUCCGAACUUGCUCUUGUUCUCGGUGCCGUCCAACUUGAUCAUGAAGUUGUCGAUGGACGCCUGGUCAACCACAUCCAGAUUGGCCUUGAUCA ...(((.....))).(((((..(((((((..((((........))))..(((((((((((((((.......))))))..))))).)))).)))))))..))))).((((......)))). (-40.12 = -40.48 + 0.36)

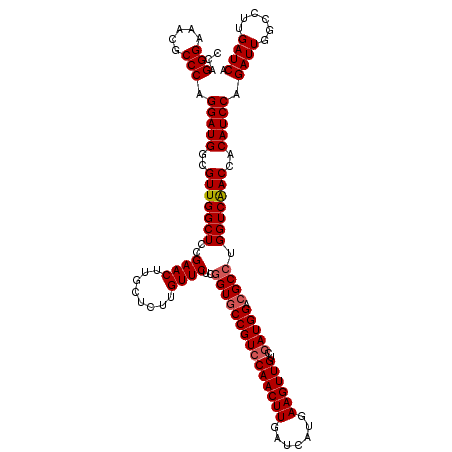
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:29 2006