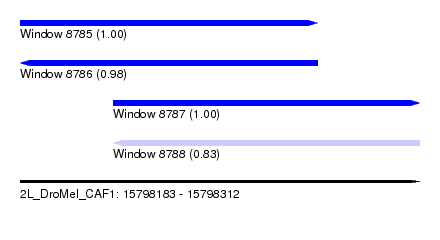
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,798,183 – 15,798,312 |
| Length | 129 |
| Max. P | 0.998909 |

| Location | 15,798,183 – 15,798,279 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -25.66 |
| Energy contribution | -25.48 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.998339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15798183 96 + 22407834 GGGAAUAUGUUUUAAAUGUUUCGCAA----------AAGCAAUUUUUUUUGUUGUCUGCCGUCUGUGUGUUUGCCUUUUGGGGCCAGCAAACAAAUGGCAAA--------------AACA .(((((((.......)))))))((..----------..)).........((((...((((((.(((.((((.((((....)))).)))).))).))))))..--------------)))) ( -28.40) >DroSec_CAF1 22107 95 + 1 GGGAUUAUGUUUUAAAUGUUUCGCAA----------AAGCAAUUU-UUUUGUUGUCUGCCGUCUGUGUGUUUGCCUUUUGGGGCCAGCAAACAAAUGGCAAA--------------AACA .......((((((.........((((----------(((.....)-))))))....((((((.(((.((((.((((....)))).)))).))).))))))))--------------)))) ( -25.90) >DroAna_CAF1 21642 117 + 1 GGGAAUAUGUUUUAAAUAUUUCCAAAAAAAAAAAAAAAACAAAAU-UCCUGUUGUCUGCUGUCUG--UGUUUGCCUUUUGGGGCCAACAAACAAACGGCACAACAAAGCCCACCAAAACA ((((((((.......))))))))......................-...((((((..(((((.((--((((.((((....)))).)))..))).)))))))))))............... ( -28.50) >DroSim_CAF1 13390 95 + 1 GGGAAUAUGUUUUAAAUGUUUCGCAA----------AAGCAAUUU-UUUUGUUGUCUGCCGUCUGUGUGUUUGCCUUUUGGGGCCAGCAAACAAAUGGCAAA--------------AACA .(((((((.......)))))))((..----------..)).....-...((((...((((((.(((.((((.((((....)))).)))).))).))))))..--------------)))) ( -28.40) >consensus GGGAAUAUGUUUUAAAUGUUUCGCAA__________AAGCAAUUU_UUUUGUUGUCUGCCGUCUGUGUGUUUGCCUUUUGGGGCCAGCAAACAAAUGGCAAA______________AACA .(((((((.......)))))))...............(((((......)))))...((((((.(((.((((.((((....)))).)))).))).)))))).................... (-25.66 = -25.48 + -0.19)

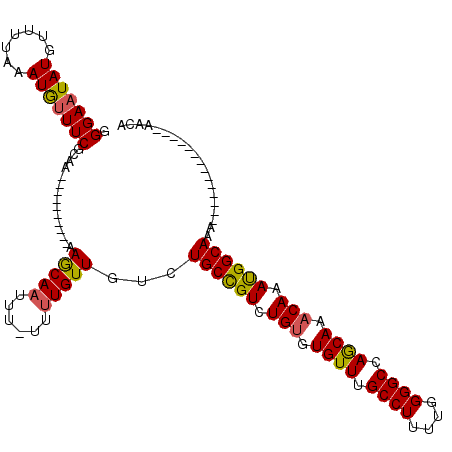

| Location | 15,798,183 – 15,798,279 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -24.68 |
| Consensus MFE | -15.09 |
| Energy contribution | -15.22 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15798183 96 - 22407834 UGUU--------------UUUGCCAUUUGUUUGCUGGCCCCAAAAGGCAAACACACAGACGGCAGACAACAAAAAAAAUUGCUU----------UUGCGAAACAUUUAAAACAUAUUCCC ((((--------------((((((.(((((.((...(((......)))...)).))))).)))))).)))).......((((..----------..)))).................... ( -23.30) >DroSec_CAF1 22107 95 - 1 UGUU--------------UUUGCCAUUUGUUUGCUGGCCCCAAAAGGCAAACACACAGACGGCAGACAACAAAA-AAAUUGCUU----------UUGCGAAACAUUUAAAACAUAAUCCC ((((--------------((((((.(((((.((...(((......)))...)).))))).)))))).))))...-...((((..----------..)))).................... ( -23.30) >DroAna_CAF1 21642 117 - 1 UGUUUUGGUGGGCUUUGUUGUGCCGUUUGUUUGUUGGCCCCAAAAGGCAAACA--CAGACAGCAGACAACAGGA-AUUUUGUUUUUUUUUUUUUUUUGGAAAUAUUUAAAACAUAUUCCC (((((.(.(((((........)))((((((((.(((....))).)))))))).--....)).))))))...(((-((..((((((.....((((....)))).....)))))).))))). ( -28.80) >DroSim_CAF1 13390 95 - 1 UGUU--------------UUUGCCAUUUGUUUGCUGGCCCCAAAAGGCAAACACACAGACGGCAGACAACAAAA-AAAUUGCUU----------UUGCGAAACAUUUAAAACAUAUUCCC ((((--------------((((((.(((((.((...(((......)))...)).))))).)))))).))))...-...((((..----------..)))).................... ( -23.30) >consensus UGUU______________UUUGCCAUUUGUUUGCUGGCCCCAAAAGGCAAACACACAGACGGCAGACAACAAAA_AAAUUGCUU__________UUGCGAAACAUUUAAAACAUAUUCCC ((((......................(((((((((((((......)))...(.....).)))))))))).........((((..............)))).........))))....... (-15.09 = -15.22 + 0.12)

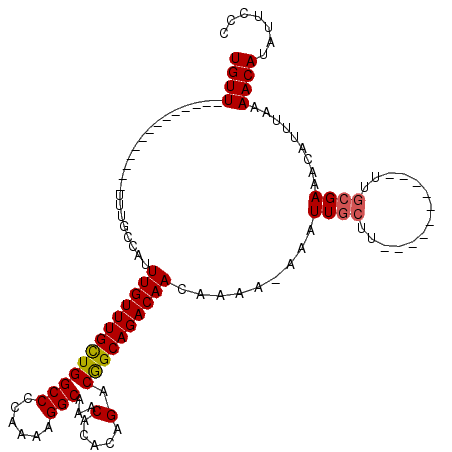
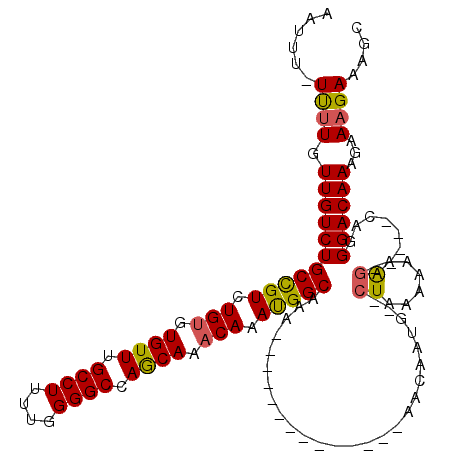
| Location | 15,798,213 – 15,798,312 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.16 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -23.26 |
| Energy contribution | -23.08 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.80 |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15798213 99 + 22407834 AAUUUUUUUUGUUGUCUGCCGUCUGUGUGUUUGCCUUUUGGGGCCAGCAAACAAAUGGCAAA--------------AACAAUG--CUAAAAAAGG-----CAGGGACAAAGAAAGAAAGC ..((((((((.(((((((((((.(((.((((.((((....)))).)))).))).)))))...--------------.....((--((......))-----)).)))))).)))))))).. ( -34.70) >DroSec_CAF1 22137 98 + 1 AAUUU-UUUUGUUGUCUGCCGUCUGUGUGUUUGCCUUUUGGGGCCAGCAAACAAAUGGCAAA--------------AACAAUG--CUAAAAAAAG-----GAGGGACAAAGAAAGAAAGC ..(((-((((.(((((((((((.(((.((((.((((....)))).)))).))).)))))...--------------.....(.--((......))-----.).)))))).)))))))... ( -29.30) >DroAna_CAF1 21682 117 + 1 AAAAU-UCCUGUUGUCUGCUGUCUG--UGUUUGCCUUUUGGGGCCAACAAACAAACGGCACAACAAAGCCCACCAAAACAACAACAUAAAAAAAGAAAUCCAGGGACAAAGAAAGAAAAC ....(-((.((((((..(((((.((--((((.((((....)))).)))..))).)))))))))))..((((...............................))).)...)))....... ( -22.98) >DroSim_CAF1 13420 99 + 1 AAUUU-UUUUGUUGUCUGCCGUCUGUGUGUUUGCCUUUUGGGGCCAGCAAACAAAUGGCAAA--------------AACAAUG--CUAAAAAAAGG----GAGGGACAAAGAAAGAAAGC ..(((-((((.(((((((((((.(((.((((.((((....)))).)))).))).)))))...--------------.....(.--((......)).----)..)))))).)))))))... ( -29.30) >consensus AAUUU_UUUUGUUGUCUGCCGUCUGUGUGUUUGCCUUUUGGGGCCAGCAAACAAAUGGCAAA______________AACAAUG__CUAAAAAAAG_____CAGGGACAAAGAAAGAAAGC ......((((.(((((((((((.(((.((((.((((....)))).)))).))).)))))..........................((......))........))))))...)))).... (-23.26 = -23.08 + -0.19)


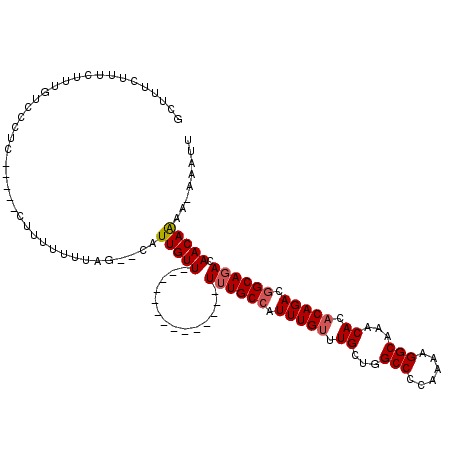
| Location | 15,798,213 – 15,798,312 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.16 |
| Mean single sequence MFE | -23.59 |
| Consensus MFE | -15.35 |
| Energy contribution | -15.91 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15798213 99 - 22407834 GCUUUCUUUCUUUGUCCCUG-----CCUUUUUUAG--CAUUGUU--------------UUUGCCAUUUGUUUGCUGGCCCCAAAAGGCAAACACACAGACGGCAGACAACAAAAAAAAUU ..................((-----(........)--))(((((--------------((((((.(((((.((...(((......)))...)).))))).)))))).)))))........ ( -22.50) >DroSec_CAF1 22137 98 - 1 GCUUUCUUUCUUUGUCCCUC-----CUUUUUUUAG--CAUUGUU--------------UUUGCCAUUUGUUUGCUGGCCCCAAAAGGCAAACACACAGACGGCAGACAACAAAA-AAAUU (((.................-----........))--).(((((--------------((((((.(((((.((...(((......)))...)).))))).)))))).)))))..-..... ( -21.91) >DroAna_CAF1 21682 117 - 1 GUUUUCUUUCUUUGUCCCUGGAUUUCUUUUUUUAUGUUGUUGUUUUGGUGGGCUUUGUUGUGCCGUUUGUUUGUUGGCCCCAAAAGGCAAACA--CAGACAGCAGACAACAGGA-AUUUU ......((((((((((..((((........))))...((((((((.(...(((........)))((((((((.(((....))).)))))))).--)))))))))))))).))))-).... ( -28.10) >DroSim_CAF1 13420 99 - 1 GCUUUCUUUCUUUGUCCCUC----CCUUUUUUUAG--CAUUGUU--------------UUUGCCAUUUGUUUGCUGGCCCCAAAAGGCAAACACACAGACGGCAGACAACAAAA-AAAUU (((.................----.........))--).(((((--------------((((((.(((((.((...(((......)))...)).))))).)))))).)))))..-..... ( -21.87) >consensus GCUUUCUUUCUUUGUCCCUC_____CUUUUUUUAG__CAUUGUU______________UUUGCCAUUUGUUUGCUGGCCCCAAAAGGCAAACACACAGACGGCAGACAACAAAA_AAAUU .......................................(((((..............((((((.(((((.((...(((......)))...)).))))).)))))).)))))........ (-15.35 = -15.91 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:38 2006