
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,795,362 – 15,795,467 |
| Length | 105 |
| Max. P | 0.851622 |

| Location | 15,795,362 – 15,795,467 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.78 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -4.43 |
| Energy contribution | -4.99 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.22 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15795362 105 + 22407834 GCUAGCUUGGACUGGAUAC-CCGCCCGCCUUGCCUU-UUCGCGAACUAAUUGCUAAAU-------------AUUUUUCAUUAAAAAUCAAAUGCAAUUUCGCUAAAAAAAAGUCUCAAGU ....(((((((((((....-))............((-((.(((((...(((((.....-------------((((((....)))))).....)))))))))).))))...)))).))))) ( -21.40) >DroSec_CAF1 19183 104 + 1 CCGAGCUUUGGCACGAUUU-CUGUGCGCCUUGCCUU-UUCGUGAACUAAUUGCUAAAU-------------AUUUUUCAUUAAAAAUCAAAUGCAAUUUCGCAAAAAAAAAG-CUCAAGU ..(((((((.(((((....-.)))))..........-...(((((...(((((.....-------------((((((....)))))).....))))))))))......))))-))).... ( -24.50) >DroAna_CAF1 18157 100 + 1 ---AGCUU-GGAAAGAUUUAAAUUGAACUUUGUUUCUUUGGCUUACAAAUUGCCAAAU-------------AUUUUCCAUUAA-AAUGAAAUGCAAUUUU--UACAAAAACCUCUCAAGU ---.((((-(((((((..((((......))))..))))).......(((((((....(-------------(((((.....))-))))....))))))).--............)))))) ( -14.10) >DroSim_CAF1 10381 100 + 1 -----CUUUGUCACGAUUU-CUGUGCGCCUUGCCUU-UUCGCGAACUAAUUGCUAAAU-------------AUUUUUCAUUAAAAAUCAAAUGCAAUUUCGCUAAAAAAAAGUCUCAAGU -----....(.((((....-.)))))..((((.(((-((.(((((...(((((.....-------------((((((....)))))).....)))))))))).....)))))...)))). ( -14.30) >DroEre_CAF1 19447 118 + 1 GCGAGCUGGGACACGAUUA-UAAAGCGCCUUUCCUUUUUCGCGAACUAAUUGCUUCAUUAAAAAUGAUUUCAUUUUUCAUUAAAAAUGAAAUGCAAUUUCGUGAA-AAAAAGUCUCAUGU ....((((((((.((....-.....)).......(((((((((((...(((((.((((.....))))((((((((((....)))))))))).)))))))))))))-)))..)))))).)) ( -28.70) >consensus _C_AGCUUGGACACGAUUU_CAGUGCGCCUUGCCUU_UUCGCGAACUAAUUGCUAAAU_____________AUUUUUCAUUAAAAAUCAAAUGCAAUUUCGCUAAAAAAAAGUCUCAAGU ........................................((((...((((((.......................................)))))))))).................. ( -4.43 = -4.99 + 0.56)

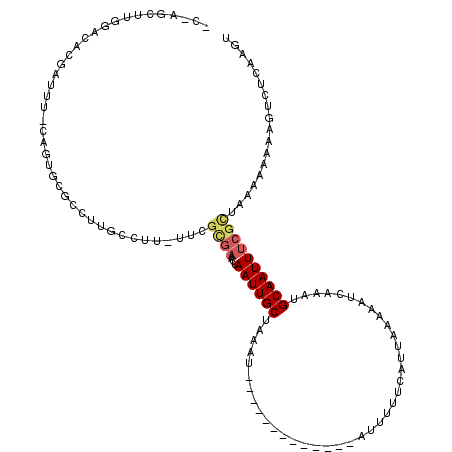
| Location | 15,795,362 – 15,795,467 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.78 |
| Mean single sequence MFE | -23.96 |
| Consensus MFE | -2.79 |
| Energy contribution | -3.59 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.12 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15795362 105 - 22407834 ACUUGAGACUUUUUUUUAGCGAAAUUGCAUUUGAUUUUUAAUGAAAAAU-------------AUUUAGCAAUUAGUUCGCGAA-AAGGCAAGGCGGGCGG-GUAUCCAGUCCAAGCUAGC .((((...((((((....((((((((((.....((((((....))))))-------------.....)))))...))))))))-))).))))..((((((-....)).))))........ ( -25.20) >DroSec_CAF1 19183 104 - 1 ACUUGAG-CUUUUUUUUUGCGAAAUUGCAUUUGAUUUUUAAUGAAAAAU-------------AUUUAGCAAUUAGUUCACGAA-AAGGCAAGGCGCACAG-AAAUCGUGCCAAAGCUCGG ...((((-(((((((((((.((((((((.....((((((....))))))-------------.....)))))...))).))))-)))....(((((....-.....))))))))))))). ( -27.70) >DroAna_CAF1 18157 100 - 1 ACUUGAGAGGUUUUUGUA--AAAAUUGCAUUUCAUU-UUAAUGGAAAAU-------------AUUUGGCAAUUUGUAAGCCAAAGAAACAAAGUUCAAUUUAAAUCUUUCC-AAGCU--- .((((((((((((.....--.((((((..((((((.-...))))))...-------------.((((((.........))))))...........)))))))))))))).)-)))..--- ( -18.20) >DroSim_CAF1 10381 100 - 1 ACUUGAGACUUUUUUUUAGCGAAAUUGCAUUUGAUUUUUAAUGAAAAAU-------------AUUUAGCAAUUAGUUCGCGAA-AAGGCAAGGCGCACAG-AAAUCGUGACAAAG----- .((((...((((((....((((((((((.....((((((....))))))-------------.....)))))...))))))))-))).))))..(((((.-...).))).)....----- ( -20.10) >DroEre_CAF1 19447 118 - 1 ACAUGAGACUUUUU-UUCACGAAAUUGCAUUUCAUUUUUAAUGAAAAAUGAAAUCAUUUUUAAUGAAGCAAUUAGUUCGCGAAAAAGGAAAGGCGCUUUA-UAAUCGUGUCCCAGCUCGC ....(((.((.(((-(((.(((((((((.((((((((((....))))))))))((((.....)))).)))))...)))).))))))((...(((((....-.....)))))))))))).. ( -28.60) >consensus ACUUGAGACUUUUUUUUAGCGAAAUUGCAUUUGAUUUUUAAUGAAAAAU_____________AUUUAGCAAUUAGUUCGCGAA_AAGGCAAGGCGCACAG_AAAUCGUGCCAAAGCU_G_ ......................((((((.......................................))))))..................(((((..........)))))......... ( -2.79 = -3.59 + 0.80)

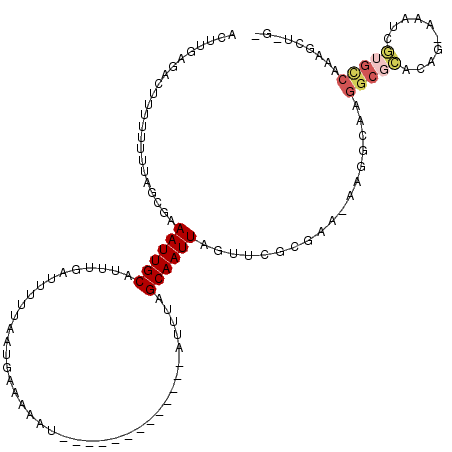
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:32 2006