
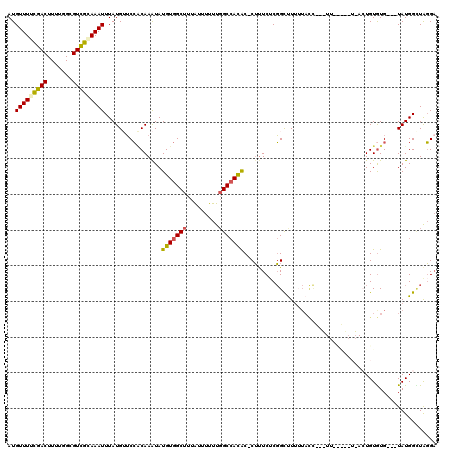
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,793,651 – 15,793,792 |
| Length | 141 |
| Max. P | 0.937242 |

| Location | 15,793,651 – 15,793,767 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.68 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -14.06 |
| Energy contribution | -14.14 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15793651 116 + 22407834 AUGUUUUCGACUUUUGGUGUCGGAAAUUUAUGUUCCACAAAUAUGUGGCUUUAUUUUUUGGCCACAC-CUUUCUCGGCUUUUUACCUUGUUCCGUAUUACUGUGUG---UAUGGAUAGGA ..(((((((((.......)))))))))................(((((((.........)))))))(-(......)).......(((...(((((((........)---)))))).))). ( -29.70) >DroSec_CAF1 17475 102 + 1 AUGUUUUCGACUUUUGGCGUCGCAAAUUUAUGUUCCACAAAUAUGUGGCUUUAUUUUUUGGCCACAC-CUUUCACGGCUUUUUACU--------------UGUGUG---UAUGGCUGGGA ..((((.((((.......)))).))))......(((.......(((((((.........))))))).-......(((((...(((.--------------...)))---...)))))))) ( -25.20) >DroAna_CAF1 16367 113 + 1 AUGUUUUUGACUUUUGGUGUUGAAAAUUUAUGGGCGGCAAAUAUGUGGCUUUGCUUUUUUGCAAUGCUCGCUCUCCGUUUA-------UUUUCAGCUAACUGUGUGACAUAUGGUUAUGA ..................(((((((((..(((((.(((........(((.((((......)))).))).))).)))))..)-------))))))))(((((((((...)))))))))... ( -28.20) >DroSim_CAF1 8666 101 + 1 AUGUUUUCGACUUUUGGCGUCGGAAAUUUAUGUUCCACAAAUAUGUGGCUUUAUUUUUUGGCCACAC-CUUUCUCGGCUUUUUACU--------------UGUGUG---UAUGGCUGGG- ..(((((((((.......)))))))))................(((((((.........))))))).-....(((((((...(((.--------------...)))---...)))))))- ( -28.50) >DroEre_CAF1 17994 116 + 1 AUGUUUUCGACUUUUGGCGUCGCAAAUUUAUGGUCCCCAAGUAUGUGGCUUUAUUUUUUGGCCACAC-CUUUCUCGGCCUUGUAUCUCUUUCUAUAUAACUGUAUG---UAUGGUUAGGA ..((((.((((.......)))).))))....((((...(((..(((((((.........))))))).-.)))...)))).......(((..(((((((......))---)))))..))). ( -25.60) >consensus AUGUUUUCGACUUUUGGCGUCGCAAAUUUAUGUUCCACAAAUAUGUGGCUUUAUUUUUUGGCCACAC_CUUUCUCGGCUUUUUACC___UU_____U_ACUGUGUG___UAUGGCUAGGA ..(((((((((.......)))))))))................(((((((.........)))))))...................................................... (-14.06 = -14.14 + 0.08)


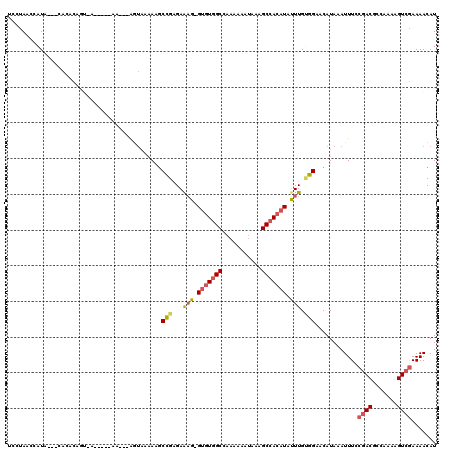
| Location | 15,793,651 – 15,793,767 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.68 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -12.70 |
| Energy contribution | -13.34 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15793651 116 - 22407834 UCCUAUCCAUA---CACACAGUAAUACGGAACAAGGUAAAAAGCCGAGAAAG-GUGUGGCCAAAAAAUAAAGCCACAUAUUUGUGGAACAUAAAUUUCCGACACCAAAAGUCGAAAACAU .....((((((---....................(((.....))).......-(((((((...........)))))))...))))))...........((((.......))))....... ( -22.50) >DroSec_CAF1 17475 102 - 1 UCCCAGCCAUA---CACACA--------------AGUAAAAAGCCGUGAAAG-GUGUGGCCAAAAAAUAAAGCCACAUAUUUGUGGAACAUAAAUUUGCGACGCCAAAAGUCGAAAACAU .....((((((---(.(((.--------------.((.....)).)))....-)))))))((((........(((((....)))))........))))((((.......))))....... ( -23.39) >DroAna_CAF1 16367 113 - 1 UCAUAACCAUAUGUCACACAGUUAGCUGAAAA-------UAAACGGAGAGCGAGCAUUGCAAAAAAGCAAAGCCACAUAUUUGCCGCCCAUAAAUUUUCAACACCAAAAGUCAAAAACAU ........................(.((((((-------(....((.(.((((((.((((......)))).)).......))))).)).....))))))).).................. ( -14.21) >DroSim_CAF1 8666 101 - 1 -CCCAGCCAUA---CACACA--------------AGUAAAAAGCCGAGAAAG-GUGUGGCCAAAAAAUAAAGCCACAUAUUUGUGGAACAUAAAUUUCCGACGCCAAAAGUCGAAAACAU -..........---..((((--------------((((....(((......)-))(((((...........))))).)))))))).............((((.......))))....... ( -23.30) >DroEre_CAF1 17994 116 - 1 UCCUAACCAUA---CAUACAGUUAUAUAGAAAGAGAUACAAGGCCGAGAAAG-GUGUGGCCAAAAAAUAAAGCCACAUACUUGGGGACCAUAAAUUUGCGACGCCAAAAGUCGAAAACAU (((((((....---......))))...................(((((....-(((((((...........))))))).))))))))...........((((.......))))....... ( -21.60) >consensus UCCUAACCAUA___CACACAGU_A_____AA___AGUAAAAAGCCGAGAAAG_GUGUGGCCAAAAAAUAAAGCCACAUAUUUGUGGAACAUAAAUUUCCGACGCCAAAAGUCGAAAACAU ...........................................(((...(((.(((((((...........))))))).))).)))............((((.......))))....... (-12.70 = -13.34 + 0.64)


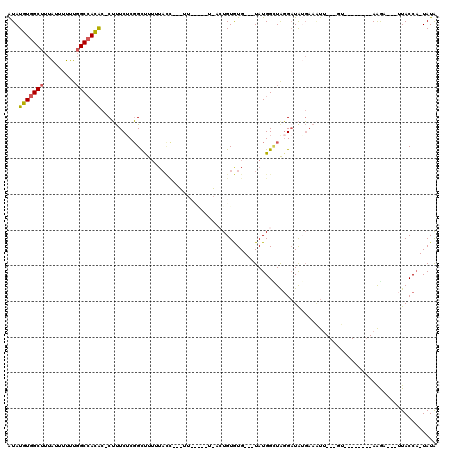
| Location | 15,793,691 – 15,793,792 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.97 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -7.52 |
| Energy contribution | -7.60 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15793691 101 + 22407834 AUAUGUGGCUUUAUUUUUUGGCCACAC-CUUUCUCGGCUUUUUACCUUGUUCCGUAUUACUGUGUG---UAUGGAUAGGAUAUGAAAUU---GU--------AAGA---UUACCA-UAUA ...(((((((.........)))))))(-(......)).......(((...(((((((........)---)))))).)))(((((.((((---..--------..))---))..))-))). ( -22.50) >DroSec_CAF1 17515 87 + 1 AUAUGUGGCUUUAUUUUUUGGCCACAC-CUUUCACGGCUUUUUACU--------------UGUGUG---UAUGGCUGGGAUAUGAAAUG---GU--------AAGA---UUACCA-UAUA ...(((((((.........))))))).-..(((.(((((...(((.--------------...)))---...))))))))......(((---((--------(...---.)))))-)... ( -24.30) >DroAna_CAF1 16407 113 + 1 AUAUGUGGCUUUGCUUUUUUGCAAUGCUCGCUCUCCGUUUA-------UUUUCAGCUAACUGUGUGACAUAUGGUUAUGAUGAUAAGUUUUCGAAGGCUCUAAGGAAUUUUACCUUUAUG ....(((((.((((......)))).)).)))..(((..(((-------((.(((..(((((((((...)))))))))))).)))))((((....)))).....))).............. ( -20.90) >DroSim_CAF1 8706 86 + 1 AUAUGUGGCUUUAUUUUUUGGCCACAC-CUUUCUCGGCUUUUUACU--------------UGUGUG---UAUGGCUGGG-UAUGAAAUU---GU--------AAGA---UUACCA-UAUA ...(((((((.........))))))).-....(((((((...(((.--------------...)))---...)))))))-((((.((((---..--------..))---))..))-)).. ( -21.10) >DroEre_CAF1 18034 104 + 1 GUAUGUGGCUUUAUUUUUUGGCCACAC-CUUUCUCGGCCUUGUAUCUCUUUCUAUAUAACUGUAUG---UAUGGUUAGGAUAAGAAUUU---GA--------AAAAAAUGUACCA-UAUA .(((((((...((((((((((((....-.......))))...((((.((..(((((((......))---)))))..)))))).......---..--------))))))))..)))-)))) ( -25.60) >consensus AUAUGUGGCUUUAUUUUUUGGCCACAC_CUUUCUCGGCUUUUUACC___UU_____U_ACUGUGUG___UAUGGCUAGGAUAUGAAAUU___GU________AAGA___UUACCA_UAUA ...(((((((.........))))))).............................................................................................. ( -7.52 = -7.60 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:27 2006