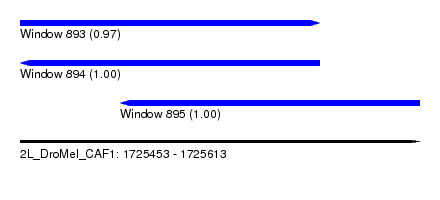
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,725,453 – 1,725,613 |
| Length | 160 |
| Max. P | 0.999749 |

| Location | 1,725,453 – 1,725,573 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -53.80 |
| Consensus MFE | -50.82 |
| Energy contribution | -51.14 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1725453 120 + 22407834 CGGUCUUGAUCUGGCCGGUGGACAGACCGACGACCAGGUCACCGAUGAACGAGUCCUCGGUCUCACCGGAACGGUGGGAGACCAUGGUGCCCCAGCCGUUCUUCUUGGCCAGCAGAUGGG (.((((....((((((((((...(((((((.(((..(.(((....))).)..))).)))))))))))(((((((((((..((....))..)))).)))))))....)))))).)))).). ( -56.00) >DroSec_CAF1 7606 120 + 1 CGGUCUUGAUCUGGCCGGUGGACAGACCGACGACCAGGUCACCGAUGAACGAGUCCUCGGUCUCGCCGGAACGGUGGGAGACCAUGGUGCCCCAGCCGUUCUUCUUGGCCAGCAGAUGGG (.((((....((((((((((...(((((((.(((..(.(((....))).)..))).)))))))))))(((((((((((..((....))..)))).)))))))....)))))).)))).). ( -55.60) >DroSim_CAF1 7784 120 + 1 CGGUCUUGAUCUGGCCGGUGGACAGACCGACGACCAAGUCACCGAUGAACGAGUCCUCGGUCUCGCCGGAACGGUGGGAGACCAUGGUGCCCCAGCCGUUCUUCUUGGCCAGCAGAUGGG (.((((....((((((((((...(((((((.(((....(((....)))....))).)))))))))))(((((((((((..((....))..)))).)))))))....)))))).)))).). ( -55.00) >DroEre_CAF1 7632 120 + 1 CGGUCUUGAUCUGGCCGGUGGACAGGCCCACGACCAGGUCACCGAUGAACGAGUCCUCGGUCUCGCCGGAGCGGUGGGAGACCAUGGUGCCCCAACCGUUCUGCUUGGCCAGCAGAUGGG (.((((....(((((((((((((.((((........))))..(((.((.....)).)))))).))))(((((((((((..((....))..))).))))))))....)))))).)))).). ( -52.10) >DroYak_CAF1 8290 120 + 1 CGGUCUUGAUCUGGCCGGUGGACAGACCCACGACCAGGUCACCGAUGAACGAGUCCUCGGUCUCGCCGGAACGGUGAGAGACCAUGGUACCCCAGCCGUUCUGCUUGGCCAGCAGAUGAG .((((.((((((((.((.(((......))))).))))))))((((.((.....)).))))((((((((...))))))))))))(((((......)))))((((((.....)))))).... ( -50.30) >consensus CGGUCUUGAUCUGGCCGGUGGACAGACCGACGACCAGGUCACCGAUGAACGAGUCCUCGGUCUCGCCGGAACGGUGGGAGACCAUGGUGCCCCAGCCGUUCUUCUUGGCCAGCAGAUGGG (.((((....((((((((((...(((((((.(((....(((....)))....))).)))))))))))(((((((((((..((....))..)))).)))))))....)))))).)))).). (-50.82 = -51.14 + 0.32)

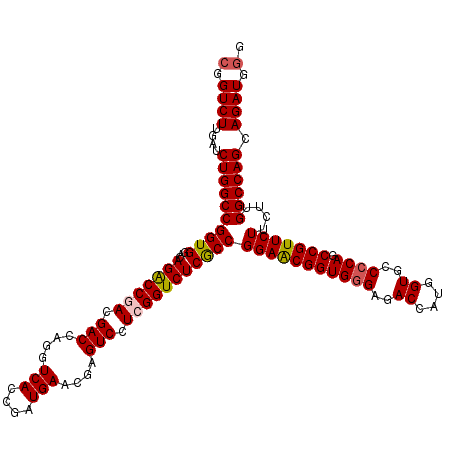
| Location | 1,725,453 – 1,725,573 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -54.66 |
| Consensus MFE | -51.70 |
| Energy contribution | -52.18 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.00 |
| SVM RNA-class probability | 0.999749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1725453 120 - 22407834 CCCAUCUGCUGGCCAAGAAGAACGGCUGGGGCACCAUGGUCUCCCACCGUUCCGGUGAGACCGAGGACUCGUUCAUCGGUGACCUGGUCGUCGGUCUGUCCACCGGCCAGAUCAAGACCG ....(((.((((((.....((((((.(((((.((....)).))))))))))).(((((((((((.((((.(.(((....))).).)))).)))))))...))))))))))....)))... ( -58.90) >DroSec_CAF1 7606 120 - 1 CCCAUCUGCUGGCCAAGAAGAACGGCUGGGGCACCAUGGUCUCCCACCGUUCCGGCGAGACCGAGGACUCGUUCAUCGGUGACCUGGUCGUCGGUCUGUCCACCGGCCAGAUCAAGACCG ....(((.((((((.....((((((.(((((.((....)).))))))))))).(((.(((((((.((((.(.(((....))).).)))).))))))))))....))))))....)))... ( -56.40) >DroSim_CAF1 7784 120 - 1 CCCAUCUGCUGGCCAAGAAGAACGGCUGGGGCACCAUGGUCUCCCACCGUUCCGGCGAGACCGAGGACUCGUUCAUCGGUGACUUGGUCGUCGGUCUGUCCACCGGCCAGAUCAAGACCG ....(((.((((((.....((((((.(((((.((....)).))))))))))).(((.(((((((.((((.(.(((....))).).)))).))))))))))....))))))....)))... ( -55.20) >DroEre_CAF1 7632 120 - 1 CCCAUCUGCUGGCCAAGCAGAACGGUUGGGGCACCAUGGUCUCCCACCGCUCCGGCGAGACCGAGGACUCGUUCAUCGGUGACCUGGUCGUGGGCCUGUCCACCGGCCAGAUCAAGACCG ....((((((.....)))))).(((((..((((((..((((((((........)).))))))(((((....))).))))))..(((((((((((....)))).))))))).))..))))) ( -52.40) >DroYak_CAF1 8290 120 - 1 CUCAUCUGCUGGCCAAGCAGAACGGCUGGGGUACCAUGGUCUCUCACCGUUCCGGCGAGACCGAGGACUCGUUCAUCGGUGACCUGGUCGUGGGUCUGUCCACCGGCCAGAUCAAGACCG ....((((((.....))))))..((((((((..(((((..(..((((((....((((((.(....).))))))...))))))...)..)))))......)).))))))............ ( -50.40) >consensus CCCAUCUGCUGGCCAAGAAGAACGGCUGGGGCACCAUGGUCUCCCACCGUUCCGGCGAGACCGAGGACUCGUUCAUCGGUGACCUGGUCGUCGGUCUGUCCACCGGCCAGAUCAAGACCG ....(((.((((((.....((((((.(((((.((....)).))))))))))).((..(((((((.((((.(.(((....))).).)))).)))))))..))...))))))....)))... (-51.70 = -52.18 + 0.48)

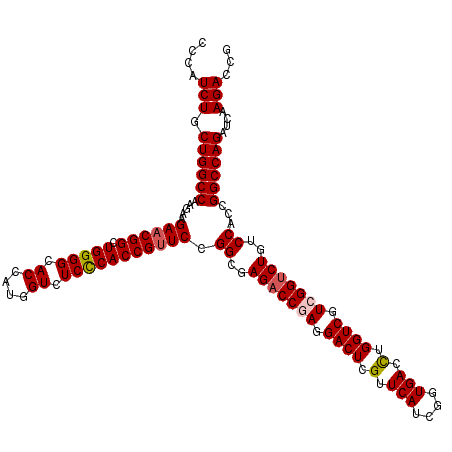
| Location | 1,725,493 – 1,725,613 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -49.86 |
| Consensus MFE | -47.56 |
| Energy contribution | -47.52 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.51 |
| SVM RNA-class probability | 0.999321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1725493 120 - 22407834 AAGGUCAACCAGAUCGGCACCGUCACCGAGUCCAUUGCCGCCCAUCUGCUGGCCAAGAAGAACGGCUGGGGCACCAUGGUCUCCCACCGUUCCGGUGAGACCGAGGACUCGUUCAUCGGU ..((((.....))))...((((....(((((((......((((....((((..(.....)..))))..))))....(((((((..((((...))))))))))).))))))).....)))) ( -48.40) >DroSec_CAF1 7646 120 - 1 AAGGUCAACCAGAUCGGCACCGUGACCGAGUCCAUUGCCGCCCAUCUGCUGGCCAAGAAGAACGGCUGGGGCACCAUGGUCUCCCACCGUUCCGGCGAGACCGAGGACUCGUUCAUCGGU ..((((.....))))...(((((((.(((((((......((((....((((..(.....)..))))..))))....(((((((((........)).))))))).))))))).))).)))) ( -52.20) >DroSim_CAF1 7824 120 - 1 AAGGUCAACCAGAUCGGCACCGUGACCGAAUCCAUUGCCGCCCAUCUGCUGGCCAAGAAGAACGGCUGGGGCACCAUGGUCUCCCACCGUUCCGGCGAGACCGAGGACUCGUUCAUCGGU ..((((.....))))...(((((((.(((.(((......((((....((((..(.....)..))))..))))....(((((((((........)).))))))).))).))).))).)))) ( -46.20) >DroEre_CAF1 7672 120 - 1 AAGGUCAACCAGAUCGGCACCGUGACCGAGUCCAUUGCCGCCCAUCUGCUGGCCAAGCAGAACGGUUGGGGCACCAUGGUCUCCCACCGCUCCGGCGAGACCGAGGACUCGUUCAUCGGU ..((((.....))))...(((((((.(((((((..(((((((..((((((.....))))))..)))...))))...(((((((((........)).))))))).))))))).))).)))) ( -54.00) >DroYak_CAF1 8330 120 - 1 AAGGUCAACCAGAUCGGUACCGUAACCGAGUCCAUUGCCGCUCAUCUGCUGGCCAAGCAGAACGGCUGGGGUACCAUGGUCUCUCACCGUUCCGGCGAGACCGAGGACUCGUUCAUCGGU ..((((.....))))...((((....(((((((...((((....((((((.....)))))).))))(((....)))(((((((...(((...))).))))))).))))))).....)))) ( -48.50) >consensus AAGGUCAACCAGAUCGGCACCGUGACCGAGUCCAUUGCCGCCCAUCUGCUGGCCAAGAAGAACGGCUGGGGCACCAUGGUCUCCCACCGUUCCGGCGAGACCGAGGACUCGUUCAUCGGU ..((((.....))))...(((((((.(((((((......((((....((((..(.....)..))))..))))....(((((((((........)).))))))).))))))).))).)))) (-47.56 = -47.52 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:12 2006