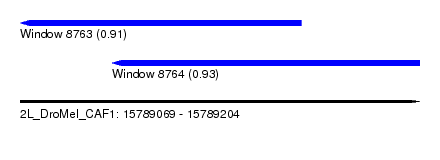
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,789,069 – 15,789,204 |
| Length | 135 |
| Max. P | 0.932666 |

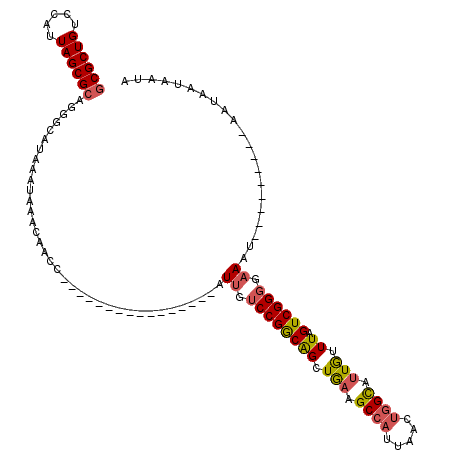
| Location | 15,789,069 – 15,789,164 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -18.58 |
| Energy contribution | -18.98 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15789069 95 - 22407834 GCGCUGUCCAUUAGCGCAGGGCAUAAAUAAACAACC----------------AUUGUCCGGCAGCUGAAGCCAUUAACUGGCAUUGUUUAGUCGGGGAAAU---------AAUAAUAAUA ((((((.....)))))).(((((.............----------------..)))))(((.......))).((..(((((........)))))..))..---------.......... ( -25.06) >DroSec_CAF1 12884 94 - 1 GCGCUGUCCAUUAGCGCCGGGCAUAAAUAAACAACC----------------AUUGUCCGGCAGCUGA-GCCAUUAACUGGCAUUGUUUAGUCGGGGAAAU---------AAUAAUAAUA .(((((..(((((((((((((((.............----------------..)))))))).)))))-((((.....))))..))..))).)).......---------.......... ( -29.66) >DroAna_CAF1 12070 120 - 1 GCGCUGUCCAUUAGCGGAUGGCGUAAAUAAACAACCAGAACCACUGGACCACAUUGUCCGACGGCUAAGGCCAUUAGCCAGUCCUAUUUAGUCGGGGCAGUAACAAUAGUAAUAAUCAUA (((((((((......)))))))))..........((((.....))))...(((((((...((((((((.....)))))).(((((........))))).)).))))).)).......... ( -36.40) >DroSim_CAF1 4054 95 - 1 GCGCUGUCCAUUAGCGCCGGGCAUAAAUAAACAACC----------------AUUGUCCGGCAGCUGAAGCCAUUAACUGGCAUUGUUUAGUCGGGGAAAU---------AAUAAUAAUA .(((((..(((((((((((((((.............----------------..)))))))).))))).((((.....))))..))..))).)).......---------.......... ( -29.66) >DroEre_CAF1 10871 95 - 1 GCGCUGUCCAUUAGCGCAGGGCAUAAAUAAACAACC----------------AUUGUCCGGCAGCUGAAGCCAUUAACUGGCUUUGUUUAGUCGGUGAAAU---------AAUAAUAAUA ((((((.....)))))).(((((.............----------------..)))))(((.......)))....(((((((......))))))).....---------.......... ( -26.96) >consensus GCGCUGUCCAUUAGCGCAGGGCAUAAAUAAACAACC________________AUUGUCCGGCAGCUGAAGCCAUUAACUGGCAUUGUUUAGUCGGGGAAAU_________AAUAAUAAUA ((((((.....))))))....................................((.((((((((.(((.((((.....)))).))).)).)))))).))..................... (-18.58 = -18.98 + 0.40)

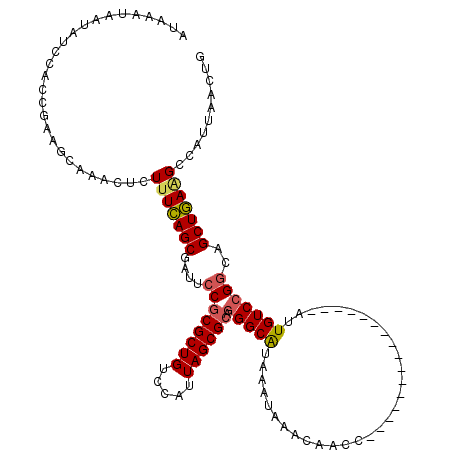
| Location | 15,789,100 – 15,789,204 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.95 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -20.22 |
| Energy contribution | -20.42 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15789100 104 - 22407834 AUAAAUAAUAUCCACCGAAGCAAACUCUUUCAGCGAUUCCGCGCUGUCCAUUAGCGCAGGGCAUAAAUAAACAACC----------------AUUGUCCGGCAGCUGAAGCCAUUAACUG ...........................(((((((....((((((((.....)))))).(((((.............----------------..)))))))..))))))).......... ( -25.66) >DroSec_CAF1 12915 103 - 1 AUAAAUAAUAUCCACCGAAGCAAACUCUUUCAGCGAUUCCGCGCUGUCCAUUAGCGCCGGGCAUAAAUAAACAACC----------------AUUGUCCGGCAGCUGA-GCCAUUAACUG ...................((.........(((((......)))))....(((((((((((((.............----------------..)))))))).)))))-))......... ( -27.96) >DroAna_CAF1 12110 120 - 1 AUAAAUAAUAUCAACCGAAGGAAACUCUUUUAGCAAUUCCGCGCUGUCCAUUAGCGGAUGGCGUAAAUAAACAACCAGAACCACUGGACCACAUUGUCCGACGGCUAAGGCCAUUAGCCA ...............((.((....))......(((((...(((((((((......)))))))))..........((((.....)))).....))))).))..((((((.....)))))). ( -31.10) >DroSim_CAF1 4085 104 - 1 AUAAAUAAUAUCCACCGAAGCAAACUCUUUCAGCGAUUCCGCGCUGUCCAUUAGCGCCGGGCAUAAAUAAACAACC----------------AUUGUCCGGCAGCUGAAGCCAUUAACUG ...................((.........(((((......)))))....(((((((((((((.............----------------..)))))))).))))).))......... ( -27.96) >DroEre_CAF1 10902 104 - 1 AUAAAUAAUAUCCACCAAAGCAAACUCUUUCAGCGAUUCCGCGCUGUCCAUUAGCGCAGGGCAUAAAUAAACAACC----------------AUUGUCCGGCAGCUGAAGCCAUUAACUG ...........................(((((((....((((((((.....)))))).(((((.............----------------..)))))))..))))))).......... ( -25.66) >consensus AUAAAUAAUAUCCACCGAAGCAAACUCUUUCAGCGAUUCCGCGCUGUCCAUUAGCGCAGGGCAUAAAUAAACAACC________________AUUGUCCGGCAGCUGAAGCCAUUAACUG ...........................(((((((....((((((((.....)))))).(((((...............................)))))))..))))))).......... (-20.22 = -20.42 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:15 2006