| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,708,430 – 1,708,547 |
| Length | 117 |
| Max. P | 0.841331 |

| Location | 1,708,430 – 1,708,547 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.79 |
| Mean single sequence MFE | -35.03 |
| Consensus MFE | -25.16 |
| Energy contribution | -25.33 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.54 |
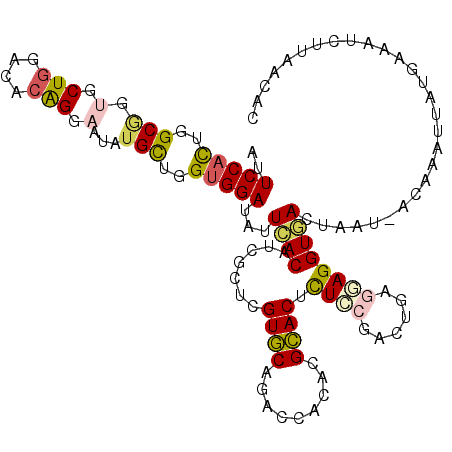
| Structure conservation index | 0.72 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
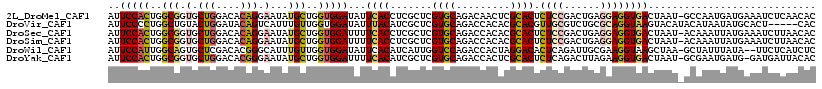
>2L_DroMel_CAF1 1708430 117 - 22407834 AUUCCACUGGCGGUGCUGGACACAGGAAUAUGCUGGUGGAUAUUCACCUCGCUCGUGCAGACAACUCGCACUCUCCGACUGAGGAGGUGACUAAU-GCCAAUGAUGAAAUCUCAACAC ..(((((..(((.(.(((....))).)...)))..)))))...(((((((....((((((....)).)))).(((.....)))))))))).....-.....((.(((....))).)). ( -38.10) >DroVir_CAF1 3326 113 - 1 AUUCCCCUGGCUGUACUGGAUACAGUCAUUUGUUGGUGGAUAUUUACAUCGCUCGUGCAGACCACACGCACGUUGCGUCUGCGCAGGUAAGUACAUACAUAAUAUGCACU-----CAC .......((((((((.....))))))))...((.((((.((((((((.(..(..((((((((((.((....)))).))))))))..)..)))).......))))).))))-----.)) ( -33.01) >DroSec_CAF1 3081 117 - 1 AUUCCACUGGCGGUGCUGGACACAGGAAUAUGCUGGUGGAUUUUCACCUCGCUCGUGCAGACCACACGCACUCUCCGACUGAGGAGGUGACUAAU-ACAAAUUAUGAAAUCUUAACAC ..(((((..(((.(.(((....))).)...)))..)))))...(((((((((..(((.....)))..)).(((.......)))))))))).....-...................... ( -36.70) >DroSim_CAF1 9897 117 - 1 AUUCCACUGGCGGUGCUGGACACAGGAAUAUGCUGGUGGAUUUUCACCUCGCUCGUGCAGACCACACGCACUCUCCGACUGAGGAGGUGACUAAU-ACAAAUUAUGAAAUCUUAACAC ..(((((..(((.(.(((....))).)...)))..)))))...(((((((((..(((.....)))..)).(((.......)))))))))).....-...................... ( -36.70) >DroWil_CAF1 5494 115 - 1 AUUCCAUUGGCAGUGCUCGACACGGGCAUUUGUUGGUGGAUAUUCACAUCAUUGGUCCAGACCACUAGGACACUCAGAUUGCGAAGGUAAGCUAA-GCUAUUUAUA--UUCUCAUCUC ..(((((..((((((((((...)))))).))))..)))))..............((((((....)).)))).((.((.((((....)))).)).)-).........--.......... ( -31.70) >DroYak_CAF1 8711 116 - 1 AUUCCACUGGCGGUGCUGGACACGGGAAUAUGCUGGUGGAUUUUCACAUCGCUCGUGCAGACCACUCGCACUCUCAGACUUAGAAGGUGACUAAU-GCGAAUGAUG-GAUGAUUACAC ..(((((..(((.(.(((....))).)...)))..)))))..((((((((((..((((((....)).)))).............((....))...-))).))).))-))......... ( -34.00) >consensus AUUCCACUGGCGGUGCUGGACACAGGAAUAUGCUGGUGGAUAUUCACAUCGCUCGUGCAGACCACACGCACUCUCCGACUGAGGAGGUGACUAAU_ACAAAUUAUGAAAUCUUAACAC ..(((((..(((.(.(((....))).)...)))..)))))...((((.......((((.........)))).((((......))))))))............................ (-25.16 = -25.33 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:09 2006