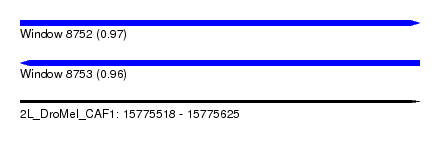
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,775,518 – 15,775,625 |
| Length | 107 |
| Max. P | 0.968541 |

| Location | 15,775,518 – 15,775,625 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.86 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -28.70 |
| Energy contribution | -29.37 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
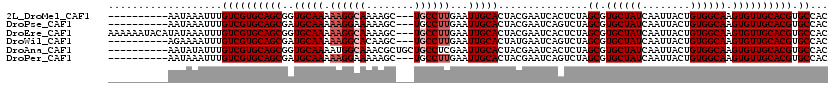
>2L_DroMel_CAF1 15775518 107 + 22407834 ----------AAUAAAUUUGUCGUGCAGCGGUGCAAAAAGGCAAAAGC---UGCCUUGAAUUGCACUACGAAUCACUCUAGCGUGCUAUCAAUUACUGUGGCAAGUGUUGCACGUGCCAC ----------.........(((((((((((((((((.((((((.....---))))))...))))))).............((.((((((........)))))).)))))))))).))... ( -36.70) >DroPse_CAF1 54500 107 + 1 ----------AAUAAAUUUGUCGUGCAGCGAUGCAAAAAGGAAAAAGC---UGCCUUGAAUUGCACUACGAAUCAGUCUAGCGUGCUAUCAAUUACUGUGGCAAGUGUUGCACGUGCCAC ----------.........((((((((((..(((((.((((.......---..))))...)))))...............((.((((((........)))))).)))))))))).))... ( -26.70) >DroEre_CAF1 28862 117 + 1 AAAAAAUACAUAUAAAUUUGUCGUGCAGCGGUGCAAAAAGGCAAAAGC---UGCCUUGAAUUGCACUACGAAUCACUCUAGCGUGCUAUCAAUUACUGUGGCAAGUGUUGCACGUGCCAC ...................(((((((((((((((((.((((((.....---))))))...))))))).............((.((((((........)))))).)))))))))).))... ( -36.70) >DroWil_CAF1 66795 107 + 1 ----------AGAAAAUUUGUCGUGCAGCGAUGCAAAAAGGCACAAGC---UGCCUUGAAUUGCACUAUGAAUCAGUCUAGCGUGCUAUCAAUUACUGUGGCAAGUGUUGCACGUGCCAC ----------.........((((((((((..(((((.((((((.....---))))))...)))))...............((.((((((........)))))).)))))))))).))... ( -31.90) >DroAna_CAF1 24150 110 + 1 ----------AAUAUAUUUGUCGUGCAGCGGUGCAAAAUGGCAAACGCUGCUGCCUCGAAUUGCACUACGAAUCACUCUAGCGUGCUAUCAAUUACUGUGGCAAGUGUUGCACGUGCCAC ----------.........(((((((((((((((((...((((........)))).....))))))).............((.((((((........)))))).)))))))))).))... ( -33.40) >DroPer_CAF1 68345 107 + 1 ----------AAUAAAUUUGUCGUGCAGCGAUGCAAAAAGGAAAAAGC---UGCCUUGAAUUGCACUACGAAUCAGUCUAGCGUGCUAUCAAUUACUGUGGCAAGUGUUGCACGUGCCAC ----------.........((((((((((..(((((.((((.......---..))))...)))))...............((.((((((........)))))).)))))))))).))... ( -26.70) >consensus __________AAUAAAUUUGUCGUGCAGCGAUGCAAAAAGGCAAAAGC___UGCCUUGAAUUGCACUACGAAUCACUCUAGCGUGCUAUCAAUUACUGUGGCAAGUGUUGCACGUGCCAC ...................((((((((((..(((((.((((((........))))))...)))))...............((.((((((........)))))).)))))))))).))... (-28.70 = -29.37 + 0.67)


| Location | 15,775,518 – 15,775,625 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.86 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -28.27 |
| Energy contribution | -28.38 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15775518 107 - 22407834 GUGGCACGUGCAACACUUGCCACAGUAAUUGAUAGCACGCUAGAGUGAUUCGUAGUGCAAUUCAAGGCA---GCUUUUGCCUUUUUGCACCGCUGCACGACAAAUUUAUU---------- ((((((.(((...))).)))))).((...((.((((.(((....))).......((((((...((((((---(...))))))).)))))).))))))..)).........---------- ( -34.60) >DroPse_CAF1 54500 107 - 1 GUGGCACGUGCAACACUUGCCACAGUAAUUGAUAGCACGCUAGACUGAUUCGUAGUGCAAUUCAAGGCA---GCUUUUUCCUUUUUGCAUCGCUGCACGACAAAUUUAUU---------- ((((((.(((...))).)))))).((..((((..((((....((.....))...))))...)))).(((---((.................)))))...)).........---------- ( -27.33) >DroEre_CAF1 28862 117 - 1 GUGGCACGUGCAACACUUGCCACAGUAAUUGAUAGCACGCUAGAGUGAUUCGUAGUGCAAUUCAAGGCA---GCUUUUGCCUUUUUGCACCGCUGCACGACAAAUUUAUAUGUAUUUUUU ((((((.(((...))).)))))).((...((.((((.(((....))).......((((((...((((((---(...))))))).)))))).))))))..))................... ( -34.60) >DroWil_CAF1 66795 107 - 1 GUGGCACGUGCAACACUUGCCACAGUAAUUGAUAGCACGCUAGACUGAUUCAUAGUGCAAUUCAAGGCA---GCUUGUGCCUUUUUGCAUCGCUGCACGACAAAUUUUCU---------- ((((((.(((...))).)))))).((...((.((((..((((((.....)).))))((((...((((((---.....)))))).))))...))))))..)).........---------- ( -31.30) >DroAna_CAF1 24150 110 - 1 GUGGCACGUGCAACACUUGCCACAGUAAUUGAUAGCACGCUAGAGUGAUUCGUAGUGCAAUUCGAGGCAGCAGCGUUUGCCAUUUUGCACCGCUGCACGACAAAUAUAUU---------- ((((((.(((...))).)))))).(((.(((....(((......)))..((((..(((........)))((((((..(((......))).))))))))))))).)))...---------- ( -34.30) >DroPer_CAF1 68345 107 - 1 GUGGCACGUGCAACACUUGCCACAGUAAUUGAUAGCACGCUAGACUGAUUCGUAGUGCAAUUCAAGGCA---GCUUUUUCCUUUUUGCAUCGCUGCACGACAAAUUUAUU---------- ((((((.(((...))).)))))).((..((((..((((....((.....))...))))...)))).(((---((.................)))))...)).........---------- ( -27.33) >consensus GUGGCACGUGCAACACUUGCCACAGUAAUUGAUAGCACGCUAGACUGAUUCGUAGUGCAAUUCAAGGCA___GCUUUUGCCUUUUUGCACCGCUGCACGACAAAUUUAUU__________ ((((((.(((...))).)))))).((...((.((((......((.....))...((((((...((((((........)))))).)))))).))))))..))................... (-28.27 = -28.38 + 0.11)

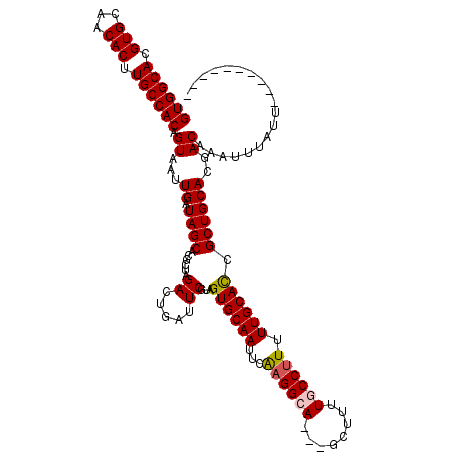

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:04 2006