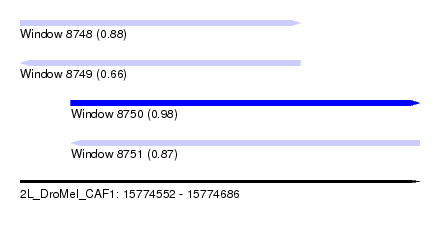
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,774,552 – 15,774,686 |
| Length | 134 |
| Max. P | 0.976131 |

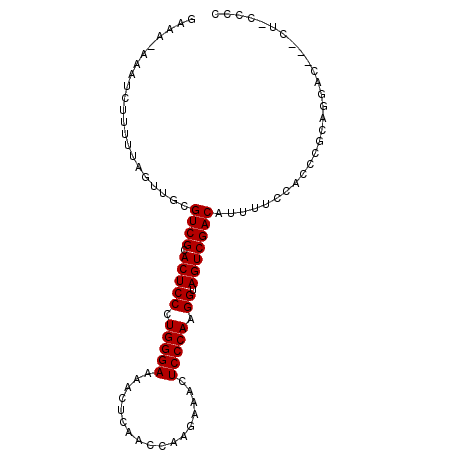
| Location | 15,774,552 – 15,774,646 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 86.37 |
| Mean single sequence MFE | -20.75 |
| Consensus MFE | -17.23 |
| Energy contribution | -17.23 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15774552 94 + 22407834 GAAA-AAAUCUUUUUAGUUGCGUCGCACUCCCUGGGAAAACUCAACCAAGGAACUCCCAAGGUAGUCGACAUUUUCCGCCCACAGGAC---CUCCCCC ....-................((((.(((((.(((((...((......))....))))).)).)))))))....(((.......))).---....... ( -20.60) >DroSec_CAF1 25866 96 + 1 GAAA-AAAUCUUUUUAGUUGCGUCGCACUCCCUGGGAAAACUUAACCAAGAAACUCCCAAGGUAGUCGACAUUUUCCACCCGCAGGACA-CCUCCCCC ....-................((((.(((((.(((((...(((....)))....))))).)).)))))))....(((.......)))..-........ ( -19.60) >DroSim_CAF1 26230 98 + 1 GAAAAAAAUCUUUUUAGUUGCGUCGCACUCCCUGGGAAAACUCAACCAAGAAACUCCCAAGGUAGUCGACAUUUUCCACCCGCAGGACAAACUACCCC ..............(((((..((((.(((((.(((((....((......))...))))).)).)))))))....(((.......)))..))))).... ( -22.30) >DroEre_CAF1 27881 86 + 1 GGAA-AAAUCUUUUUAGUUGCGUCGCACUCCCUGGGAAAACCCAACCGAGAAACUCCCAAGGUAGUCGACAUUUCCAACCCGCAGCC----------- ((((-(...((....))....((((.(((((.(((((.................))))).)).))))))).)))))...........----------- ( -20.43) >DroYak_CAF1 27334 86 + 1 GAAA-AAAUCUUUUUAGUUGCGUCGCACUCCCUGGGAAAACUCAACCAAGAAACUCCCAAGGUAGUCGACAUUUCUACCCCGCAGCC----------- ....-...........(((((((((.(((((.(((((....((......))...))))).)).)))))))...........))))).----------- ( -20.80) >consensus GAAA_AAAUCUUUUUAGUUGCGUCGCACUCCCUGGGAAAACUCAACCAAGAAACUCCCAAGGUAGUCGACAUUUUCCACCCGCAGGAC___CU_CCCC .....................((((.(((((.(((((.................))))).)).)))))))............................ (-17.23 = -17.23 + 0.00)

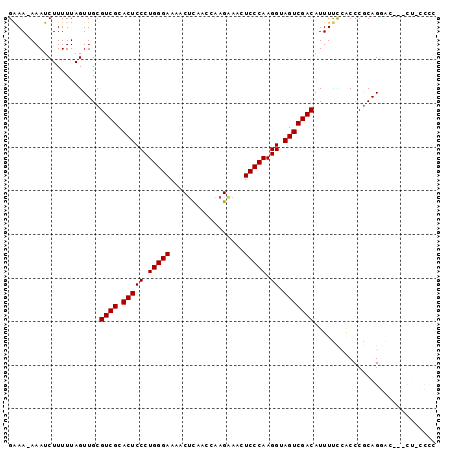
| Location | 15,774,552 – 15,774,646 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 86.37 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -25.08 |
| Energy contribution | -24.60 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.655188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15774552 94 - 22407834 GGGGGAG---GUCCUGUGGGCGGAAAAUGUCGACUACCUUGGGAGUUCCUUGGUUGAGUUUUCCCAGGGAGUGCGACGCAACUAAAAAGAUUU-UUUC .(..(((---((((((....))).....(((((((.(((((((((...(((....)))..)))))))))))).))))...........)))))-)..) ( -31.70) >DroSec_CAF1 25866 96 - 1 GGGGGAGG-UGUCCUGCGGGUGGAAAAUGUCGACUACCUUGGGAGUUUCUUGGUUAAGUUUUCCCAGGGAGUGCGACGCAACUAAAAAGAUUU-UUUC .(..((((-(.(((.......)))....(((((((.(((((((((...(((....)))..)))))))))))).))))............))))-)..) ( -29.50) >DroSim_CAF1 26230 98 - 1 GGGGUAGUUUGUCCUGCGGGUGGAAAAUGUCGACUACCUUGGGAGUUUCUUGGUUGAGUUUUCCCAGGGAGUGCGACGCAACUAAAAAGAUUUUUUUC (..((..((..(((...)))..))....(((((((.(((((((((.(((......)))..)))))))))))).))))))..)................ ( -29.40) >DroEre_CAF1 27881 86 - 1 -----------GGCUGCGGGUUGGAAAUGUCGACUACCUUGGGAGUUUCUCGGUUGGGUUUUCCCAGGGAGUGCGACGCAACUAAAAAGAUUU-UUCC -----------..((...(((((....((((((((.(((((((((...(((....)))..)))))))))))).))))))))))....))....-.... ( -29.80) >DroYak_CAF1 27334 86 - 1 -----------GGCUGCGGGGUAGAAAUGUCGACUACCUUGGGAGUUUCUUGGUUGAGUUUUCCCAGGGAGUGCGACGCAACUAAAAAGAUUU-UUUC -----------((.(((.....(....)(((((((.(((((((((.(((......)))..)))))))))))).))))))).))..........-.... ( -26.80) >consensus GGGG_AG___GUCCUGCGGGUGGAAAAUGUCGACUACCUUGGGAGUUUCUUGGUUGAGUUUUCCCAGGGAGUGCGACGCAACUAAAAAGAUUU_UUUC ............................(((((((.(((((((((...(((....)))..)))))))))))).))))..................... (-25.08 = -24.60 + -0.48)

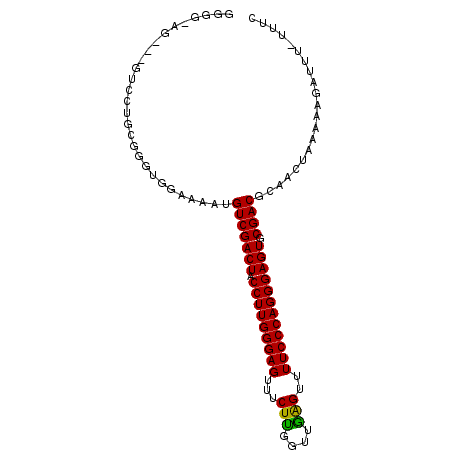
| Location | 15,774,569 – 15,774,686 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.27 |
| Mean single sequence MFE | -19.78 |
| Consensus MFE | -17.23 |
| Energy contribution | -17.23 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15774569 117 + 22407834 UGCGUCGCACUCCCUGGGAAAACUCAACCAAGGAACUCCCAAGGUAGUCGACAUUUUCCGCCCACAGGAC---CUCCCCCCCUCCGAAUCCCCCCUCCCCCGCUGAAUCCCCACCUUCCA .((((((.(((((.(((((...((......))....))))).)).)))))))....(((.......))).---............................))................. ( -22.60) >DroSec_CAF1 25883 104 + 1 UGCGUCGCACUCCCUGGGAAAACUUAACCAAGAAACUCCCAAGGUAGUCGACAUUUUCCACCCGCAGGACA-CCUCCCCCCC---------------ACCACCCGAAUCACCUCCUUCCC ...((((.(((((.(((((...(((....)))....))))).)).)))))))....(((.......)))..-..........---------------....................... ( -19.60) >DroSim_CAF1 26248 105 + 1 UGCGUCGCACUCCCUGGGAAAACUCAACCAAGAAACUCCCAAGGUAGUCGACAUUUUCCACCCGCAGGACAAACUACCCCCC---------------ACCACCCGAAUCCCCUCCUUUCC ...((((.(((((.(((((....((......))...))))).)).)))))))....(((.......))).............---------------....................... ( -19.70) >DroEre_CAF1 27898 89 + 1 UGCGUCGCACUCCCUGGGAAAACCCAACCGAGAAACUCCCAAGGUAGUCGACAUUUCCAACCCGCAGCC-----------------------------CCACACGAAUCC--ACAUUCCA ...((((.(((((.(((((.................))))).)).))))))).................-----------------------------............--........ ( -17.23) >consensus UGCGUCGCACUCCCUGGGAAAACUCAACCAAGAAACUCCCAAGGUAGUCGACAUUUUCCACCCGCAGGAC___CUCCCCCCC_______________ACCACCCGAAUCCCCACCUUCCA ...((((.(((((.(((((.................))))).)).))))))).................................................................... (-17.23 = -17.23 + 0.00)

| Location | 15,774,569 – 15,774,686 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.27 |
| Mean single sequence MFE | -35.67 |
| Consensus MFE | -25.11 |
| Energy contribution | -24.55 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15774569 117 - 22407834 UGGAAGGUGGGGAUUCAGCGGGGGAGGGGGGAUUCGGAGGGGGGGAG---GUCCUGUGGGCGGAAAAUGUCGACUACCUUGGGAGUUCCUUGGUUGAGUUUUCCCAGGGAGUGCGACGCA .....((.(..((((((((.((((((......(((.((((..(((..---..)))((.((((.....)))).))..)))).))).)))))).))))))))..))).....(((...))). ( -39.40) >DroSec_CAF1 25883 104 - 1 GGGAAGGAGGUGAUUCGGGUGGU---------------GGGGGGGAGG-UGUCCUGCGGGUGGAAAAUGUCGACUACCUUGGGAGUUUCUUGGUUAAGUUUUCCCAGGGAGUGCGACGCA .........((..(((.(.(.(.---------------.(((.(....-).)))..).).).)))...(((((((.(((((((((...(((....)))..)))))))))))).)))))). ( -34.60) >DroSim_CAF1 26248 105 - 1 GGAAAGGAGGGGAUUCGGGUGGU---------------GGGGGGUAGUUUGUCCUGCGGGUGGAAAAUGUCGACUACCUUGGGAGUUUCUUGGUUGAGUUUUCCCAGGGAGUGCGACGCA .............(((.(.(.(.---------------.(((.(.....).)))..).).).)))...(((((((.(((((((((.(((......)))..)))))))))))).))))... ( -36.20) >DroEre_CAF1 27898 89 - 1 UGGAAUGU--GGAUUCGUGUGG-----------------------------GGCUGCGGGUUGGAAAUGUCGACUACCUUGGGAGUUUCUCGGUUGGGUUUUCCCAGGGAGUGCGACGCA ..((((..--..))))((((.(-----------------------------.(((...((((((.....)))))).(((((((((...(((....)))..)))))))))))).).)))). ( -32.50) >consensus GGGAAGGAGGGGAUUCGGGUGGU_______________GGGGGGGAG___GUCCUGCGGGUGGAAAAUGUCGACUACCUUGGGAGUUUCUUGGUUGAGUUUUCCCAGGGAGUGCGACGCA ....................................................................(((((((.(((((((((...(((....)))..)))))))))))).))))... (-25.11 = -24.55 + -0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:02 2006