
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,764,022 – 15,764,178 |
| Length | 156 |
| Max. P | 0.978963 |

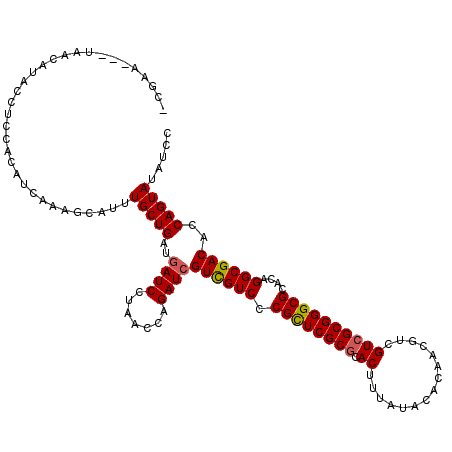
| Location | 15,764,022 – 15,764,138 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.60 |
| Mean single sequence MFE | -29.04 |
| Consensus MFE | -27.12 |
| Energy contribution | -27.12 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15764022 116 + 22407834 -CGAA---UAACAUACCUCCACAUCAAAGCAUUUGCUGAUAAUCCUAACCAGAUCGUCGUCCCGCUCGCGCACUUUGUAUACAACGUCGUCGCGGGCGCACAGGCGACACCAGUAUAUCC -....---....((((......((((..((....))))))...............((((((.(((((((((((.(((....))).)).).))))))))....))))))....)))).... ( -29.50) >DroSec_CAF1 15441 117 + 1 UUGAA---UAACAUACCUCCACAUCGAAGCAUUUGCUGAUGAUCCUAACCAGAUCGUUGUCCCGUUCGCGCACUUUAUACACAACGUCGUCGCGGGCGCACAGGCGACACCAGUAUAUCC .....---.........................(((((..((((.......))))((((((.((((((((.((...............))))))))))....))))))..)))))..... ( -25.46) >DroSim_CAF1 15634 117 + 1 AAGAA---UAACAUACCUCCACAUCAAAGCAUUUGCUGAUGAUCCUAACCAGAUCGUUGUCCCGCUCGCGCACUUUAUACACAACGUCGUCGCGGGCGCACAGGCGACACCAGUAUAUCC .....---.........................(((((..((((.......))))((((((.((((((((.((...............))))))))))....))))))..)))))..... ( -27.96) >DroEre_CAF1 15442 116 + 1 -CGAA---UAACGUACCUCUACAUCAAAGCAUUUGCUGAUGAUCUUUACCAGAUCGUCGUCCCGCUCGCUUACUUUGUAUACAACGUCGUCGCGGGCGCACAGGCGACACCAGUAUAUCC -....---....(((....)))...........(((((..(((((.....)))))((((((.(((((((..((..(((.....)))..)).)))))))....))))))..)))))..... ( -30.70) >DroYak_CAF1 15541 119 + 1 -CAAAUAAUAACCUACCUCUACAUCAAAACAUUUGCUGAUGAUCUUUACCAGAUCGUCGUCCCGCUCGCGUACUUUAUACACAACGUCGUCGCGGGCGCACAGGCGACACCAGUAUAUCC -................................(((((..(((((.....)))))((((((.((((((((.((...............))))))))))....))))))..)))))..... ( -31.56) >consensus _CGAA___UAACAUACCUCCACAUCAAAGCAUUUGCUGAUGAUCCUAACCAGAUCGUCGUCCCGCUCGCGCACUUUAUACACAACGUCGUCGCGGGCGCACAGGCGACACCAGUAUAUCC .................................(((((..((((.......))))((((((.((((((((.((...............))))))))))....))))))..)))))..... (-27.12 = -27.12 + -0.00)

| Location | 15,764,058 – 15,764,178 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.92 |
| Mean single sequence MFE | -35.66 |
| Consensus MFE | -33.60 |
| Energy contribution | -33.36 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.978963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15764058 120 + 22407834 AAUCCUAACCAGAUCGUCGUCCCGCUCGCGCACUUUGUAUACAACGUCGUCGCGGGCGCACAGGCGACACCAGUAUAUCCAAUCGCUUGUGGGAACUCCCAUGGCUAAUCCUCAACAGAA .............((((((((.(((((((((((.(((....))).)).).))))))))....))))))................(((.(((((....))))))))............)). ( -35.40) >DroSec_CAF1 15478 120 + 1 GAUCCUAACCAGAUCGUUGUCCCGUUCGCGCACUUUAUACACAACGUCGUCGCGGGCGCACAGGCGACACCAGUAUAUCCAAUCGCUCGUGGGAACUCCCAUGGCUAAUCCUCAACAGAA ((((.......))))((((((.((((((((.((...............))))))))))....))))))................((.((((((....))))))))............... ( -33.06) >DroSim_CAF1 15671 120 + 1 GAUCCUAACCAGAUCGUUGUCCCGCUCGCGCACUUUAUACACAACGUCGUCGCGGGCGCACAGGCGACACCAGUAUAUCCAAUCGCUCGUGGGAACUCCCAUGGCUAAUCCUCAAAAGAA ((((.......))))((((((.((((((((.((...............))))))))))....))))))................((.((((((....))))))))............... ( -35.56) >DroEre_CAF1 15478 120 + 1 GAUCUUUACCAGAUCGUCGUCCCGCUCGCUUACUUUGUAUACAACGUCGUCGCGGGCGCACAGGCGACACCAGUAUAUCCAAUCGCUUGUGGGUACUCCCAUGGCUAAUCCUGAAGAGAA ..(((((..(((...((((((.(((((((..((..(((.....)))..)).)))))))....))))))................(((.(((((....)))))))).....))).))))). ( -35.50) >DroYak_CAF1 15580 120 + 1 GAUCUUUACCAGAUCGUCGUCCCGCUCGCGUACUUUAUACACAACGUCGUCGCGGGCGCACAGGCGACACCAGUAUAUCCAAUCGCUCGUGGGAACUCCCAUGGCCAAUCCUCAACAGAA (((((.....)))))((((((.((((((((.((...............))))))))))....))))))................((.((((((....))))))))............... ( -38.76) >consensus GAUCCUAACCAGAUCGUCGUCCCGCUCGCGCACUUUAUACACAACGUCGUCGCGGGCGCACAGGCGACACCAGUAUAUCCAAUCGCUCGUGGGAACUCCCAUGGCUAAUCCUCAACAGAA ((((.......))))((((((.((((((((.((...............))))))))))....))))))................((.((((((....))))))))............... (-33.60 = -33.36 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:57 2006