
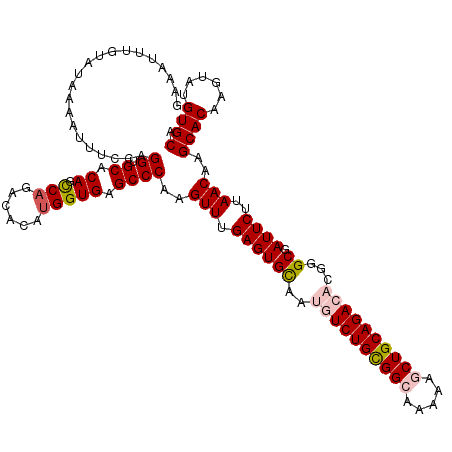
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,762,870 – 15,763,030 |
| Length | 160 |
| Max. P | 0.997206 |

| Location | 15,762,870 – 15,762,990 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -41.22 |
| Consensus MFE | -36.32 |
| Energy contribution | -37.04 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15762870 120 - 22407834 GGUGAGCCCAAGUUUGAGUGCAAUGUCUGCGGCAAAAAGCUGCAGACACGGGCGAUUCUUAACAAGCACAAGUAUGUGCACACGGAUGAGCGGCGCUUCAAGUGCGAGGUUUGUGGCAGC .(((.((((..((......))..((((((((((.....)))))))))).))))............(((((....))))).)))......((.(((((((......)))))..)).))... ( -45.10) >DroSec_CAF1 14289 120 - 1 GGUGAGCCCAAGUUUGAGUGCAAUGUCUGCGGCAAGAAGCUGCAGACACGGGCGAUUCUUAACAAGCACAAGUAUGUGCACACGGAUGAGCGGCGCUUCAAGUGCGAGGUUUGUGGCAGC .(((.((((..((......))..((((((((((.....)))))))))).))))............(((((....))))).)))......((.(((((((......)))))..)).))... ( -45.10) >DroSim_CAF1 14482 120 - 1 GGUGAGCCCAAGUUCGAGUGCAAUGUCUGCGGCAAGAAGCUGCAGACACGGGCGAUUCUUAACAAGCACAAGUAUGUGCACACGGAUGAGCGGCGCUUCAAGUGCGAGGUUUGCGGCAAC .(..(((((..((((((((((..((((((((((.....))))))))))...)).)))).......(((((....)))))........)))).((((.....))))).))))..)...... ( -47.70) >DroEre_CAF1 14302 120 - 1 GGUGAGCCCAGGUUUGAGUGCAACAUCUGUGGGAAAAAACUGCAGACUCGGGCGAUUCUUAACAAGCACAAGUACGUGCACACAGAGGAGCGACGCUUCAAGUGCGAGGUUUGUGGCAGC .(((.((((((((......))....((((..(.......)..)))))).))))............((((......)))).)))......((.(((((((......)))))..)).))... ( -34.50) >DroYak_CAF1 14408 120 - 1 GGUGAGCCCAGGUUUGAGUGUAAUAUCUGCGGCAAAAAACUGCAGACUCGGGCAAUUCUUAACAAGCACAAGUAUGUGCACACGGAGGAAAGACGCUUUAAGUGCGAGGUUUGUGGCAGC .....(((((((((((((.......(((((((.......))))))))))))))..(((((.....(((((....))))).....)))))....(((.......)))....))).)))... ( -33.71) >consensus GGUGAGCCCAAGUUUGAGUGCAAUGUCUGCGGCAAAAAGCUGCAGACACGGGCGAUUCUUAACAAGCACAAGUAUGUGCACACGGAUGAGCGGCGCUUCAAGUGCGAGGUUUGUGGCAGC .(((.((((..((......))..((((((((((.....)))))))))).))))............((((......)))).)))......((.(((((((......)))))..)).))... (-36.32 = -37.04 + 0.72)

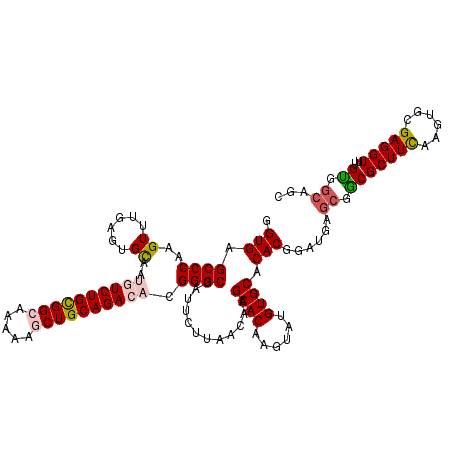

| Location | 15,762,910 – 15,763,030 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.08 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -27.66 |
| Energy contribution | -27.82 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15762910 120 + 22407834 UGCACAUACUUGUGCUUGUUAAGAAUCGCCCGUGUCUGCAGCUUUUUGCCGCAGACAUUGCACUCAAACUUGGGCUCACCAUGUGUCUGACUGUGCACCUGGAAAUUGUAUACAAAUUUC .(((((....)))))((((..(.(((..((.((((((((.((.....)).))))))))((((((((.((.(((.....)))...)).)))..)))))...))..))).)..))))..... ( -37.70) >DroSec_CAF1 14329 120 + 1 UGCACAUACUUGUGCUUGUUAAGAAUCGCCCGUGUCUGCAGCUUCUUGCCGCAGACAUUGCACUCAAACUUGGGCUCACCAUGUGUCUGGCUGUGCACCUGGAAAUUUUAUACAAAUUAC .(((((....)))))((((..(((((..((.((((((((.((.....)).))))))))((((((((.((.(((.....)))...)).)))..)))))...))..)))))..))))..... ( -38.30) >DroSim_CAF1 14522 120 + 1 UGCACAUACUUGUGCUUGUUAAGAAUCGCCCGUGUCUGCAGCUUCUUGCCGCAGACAUUGCACUCGAACUUGGGCUCACCGUGUGUCUGGCUGUGCACCUGGAAAUUUUAUACAAAUUUC .(((((....)))))((((..(((((..((.((((((((.((.....)).))))))))(((((......(..(((.(.....).)))..)..)))))...))..)))))..))))..... ( -37.00) >DroEre_CAF1 14342 120 + 1 UGCACGUACUUGUGCUUGUUAAGAAUCGCCCGAGUCUGCAGUUUUUUCCCACAGAUGUUGCACUCAAACCUGGGCUCACCAUGUGUCUGGCUGUGCACCUAGAAGUGAUAAGAGAAUUCC .(((((....))))).........(((((..((((..((((..(((......)))..))))))))....((((((.(((((......))).)).)).)).))..)))))........... ( -26.20) >DroYak_CAF1 14448 120 + 1 UGCACAUACUUGUGCUUGUUAAGAAUUGCCCGAGUCUGCAGUUUUUUGCCGCAGAUAUUACACUCAAACCUGGGCUCACCAUGUGUCUGGCUGUGCACCUAGAAAUGAUAUUAAAAUUAC ((((((..((((.......))))....(((.((((((((.((.....)).))))))...........((.(((.....))).)).)).)))))))))....................... ( -27.10) >consensus UGCACAUACUUGUGCUUGUUAAGAAUCGCCCGUGUCUGCAGCUUUUUGCCGCAGACAUUGCACUCAAACUUGGGCUCACCAUGUGUCUGGCUGUGCACCUGGAAAUUAUAUACAAAUUAC .(((((....)))))............(((((.((((((.((.....)).)))))).(((....)))...)))))...(((.((((........)))).))).................. (-27.66 = -27.82 + 0.16)

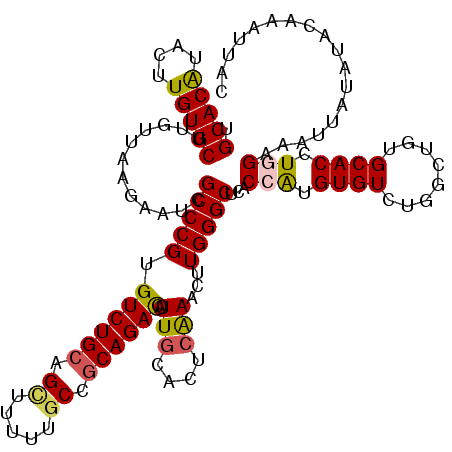
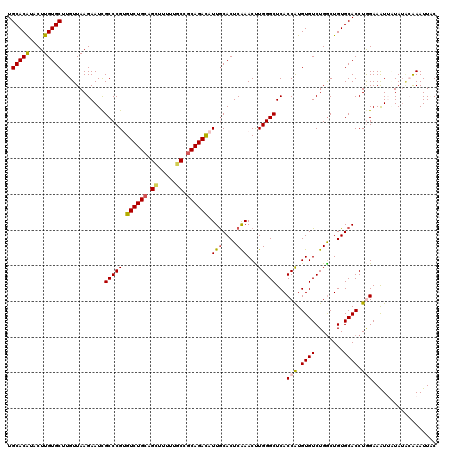
| Location | 15,762,910 – 15,763,030 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.08 |
| Mean single sequence MFE | -37.60 |
| Consensus MFE | -32.58 |
| Energy contribution | -33.50 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15762910 120 - 22407834 GAAAUUUGUAUACAAUUUCCAGGUGCACAGUCAGACACAUGGUGAGCCCAAGUUUGAGUGCAAUGUCUGCGGCAAAAAGCUGCAGACACGGGCGAUUCUUAACAAGCACAAGUAUGUGCA .....((((....((((.((...(((((..((((((...(((.....))).))))))))))).((((((((((.....))))))))))..)).))))....))))(((((....))))). ( -41.40) >DroSec_CAF1 14329 120 - 1 GUAAUUUGUAUAAAAUUUCCAGGUGCACAGCCAGACACAUGGUGAGCCCAAGUUUGAGUGCAAUGUCUGCGGCAAGAAGCUGCAGACACGGGCGAUUCUUAACAAGCACAAGUAUGUGCA .....................((.((...((((......))))..))))..(((.((((((..((((((((((.....))))))))))...)).))))..)))..(((((....))))). ( -40.50) >DroSim_CAF1 14522 120 - 1 GAAAUUUGUAUAAAAUUUCCAGGUGCACAGCCAGACACACGGUGAGCCCAAGUUCGAGUGCAAUGUCUGCGGCAAGAAGCUGCAGACACGGGCGAUUCUUAACAAGCACAAGUAUGUGCA (((((((.....)))))))..((.((...(((........)))..))))..(((.((((((..((((((((((.....))))))))))...)).))))..)))..(((((....))))). ( -41.30) >DroEre_CAF1 14342 120 - 1 GGAAUUCUCUUAUCACUUCUAGGUGCACAGCCAGACACAUGGUGAGCCCAGGUUUGAGUGCAACAUCUGUGGGAAAAAACUGCAGACUCGGGCGAUUCUUAACAAGCACAAGUACGUGCA ((((((......((((((((.(((.....)))))).....)))))((((((((......))....((((..(.......)..)))))).))))))))))......((((......)))). ( -33.50) >DroYak_CAF1 14448 120 - 1 GUAAUUUUAAUAUCAUUUCUAGGUGCACAGCCAGACACAUGGUGAGCCCAGGUUUGAGUGUAAUAUCUGCGGCAAAAAACUGCAGACUCGGGCAAUUCUUAACAAGCACAAGUAUGUGCA ............(((..(((.((.((...((((......))))..)))))))..))).(((....(((((((.......))))))).....)))...........(((((....))))). ( -31.30) >consensus GAAAUUUGUAUAAAAUUUCCAGGUGCACAGCCAGACACAUGGUGAGCCCAAGUUUGAGUGCAAUGUCUGCGGCAAAAAGCUGCAGACACGGGCGAUUCUUAACAAGCACAAGUAUGUGCA .....................((.((.((.(((......))))).))))..(((.((((((..((((((((((.....))))))))))...)).))))..)))..((((......)))). (-32.58 = -33.50 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:55 2006