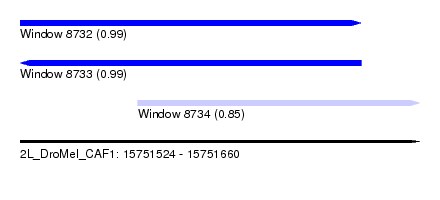
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,751,524 – 15,751,660 |
| Length | 136 |
| Max. P | 0.990757 |

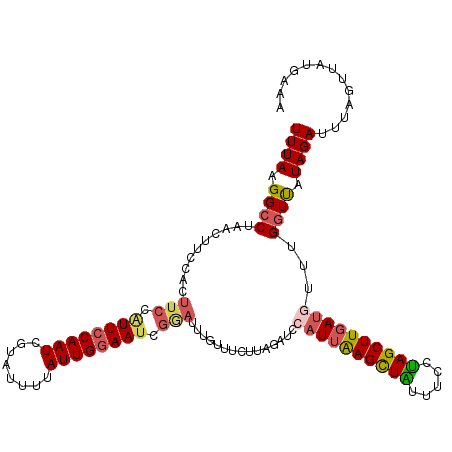
| Location | 15,751,524 – 15,751,640 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -16.71 |
| Energy contribution | -17.48 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15751524 116 + 22407834 UUUAAGGCCUAACUUCCACUUCCGUUCCAAUCGUAUUUUAUUGAAAUCGGAUUUGUUCUUUAGAUCCAUUAAGCUAUUUUCUAGCUUGAUGUUUGGUUAUAGAUUUAGUUUUGAAA ((((((((........((..((((((.((((........)))).)).))))..)).(((...((((((((((((((.....))))))))))...))))..)))....)))))))). ( -23.60) >DroSec_CAF1 3230 110 + 1 UUUAAGGCCUAACUUCCACUUCCAUUCCAAUCGUAUUUUAUUGGAAUCGGAUUUGUUUCUUAGAUCCAUUAAGUUAUUUCCUAGCUUGAUGUUUGGUCAUAGAUUUAGUU------ ((((.((((.(((..........((((((((........)))))))).(((((((.....)))))))(((((((((.....)))))))))))).)))).)))).......------ ( -27.00) >DroSim_CAF1 3229 116 + 1 UUUAAGGCCUAACUUCCACUUCCAUUCCAAUCGUAUUUUAUUGGAAUCGGAUUUGUUUCUUAGAUCCAUUAAGCUAUUUCCUAGCUUGAUGUUUGGUCAUAGAUUUAGUUUUGAAA ((((((((........((..(((((((((((........)))))))).)))..))..(((..((((((((((((((.....))))))))))...))))..)))....)))))))). ( -30.90) >DroEre_CAF1 3297 96 + 1 UUUAAGGC--------------CAUUCCAAUCGUAUUUUAUUGGAAG-AAAGUUGUUUCUAAGAUCCAUUGAGCUGUUUCCUAGCUUGAA-----GUUAUAGACUUAGCUAUAAAA ........--------------..(((((((........))))))).-..(((((..((((.(((...(..(((((.....)))))..).-----))).))))..)))))...... ( -19.90) >DroYak_CAF1 3220 116 + 1 UUUAAGGCCCAACUACCACUUCCAUUCCAAUCGUAUUUUAUUGGAAUUGAAGUUGUUUCUAAUAUCCAUUGAGCUGUUUCCCAGCUUCAUCUUUGAUUGUAGAUUUACUUAUGAGA ..((((........((.(((((.((((((((........)))))))).))))).)).((((..(((...(((((((.....)))).))).....)))..))))....))))..... ( -25.00) >consensus UUUAAGGCCUAACUUCCACUUCCAUUCCAAUCGUAUUUUAUUGGAAUCGGAUUUGUUUCUUAGAUCCAUUAAGCUAUUUCCUAGCUUGAUGUUUGGUUAUAGAUUUAGUUAUGAAA ((((.((((..........(((.((((((((........)))))))).)))...............((((((((((.....))))))))))...)))).))))............. (-16.71 = -17.48 + 0.77)

| Location | 15,751,524 – 15,751,640 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -14.28 |
| Energy contribution | -16.24 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15751524 116 - 22407834 UUUCAAAACUAAAUCUAUAACCAAACAUCAAGCUAGAAAAUAGCUUAAUGGAUCUAAAGAACAAAUCCGAUUUCAAUAAAAUACGAUUGGAACGGAAGUGGAAGUUAGGCCUUAAA .........................(((.((((((.....)))))).)))(..((((....((..((((.(((((((........)))))))))))..))....))))..)..... ( -24.80) >DroSec_CAF1 3230 110 - 1 ------AACUAAAUCUAUGACCAAACAUCAAGCUAGGAAAUAACUUAAUGGAUCUAAGAAACAAAUCCGAUUCCAAUAAAAUACGAUUGGAAUGGAAGUGGAAGUUAGGCCUUAAA ------..(((..(..(((......)))..)..)))........((((.((.(((((....((..(((.((((((((........)))))))))))..))....))))))))))). ( -19.80) >DroSim_CAF1 3229 116 - 1 UUUCAAAACUAAAUCUAUGACCAAACAUCAAGCUAGGAAAUAGCUUAAUGGAUCUAAGAAACAAAUCCGAUUCCAAUAAAAUACGAUUGGAAUGGAAGUGGAAGUUAGGCCUUAAA ........((((.(((((.......(((.((((((.....)))))).)))...............(((.((((((((........))))))))))).)))))..))))........ ( -25.80) >DroEre_CAF1 3297 96 - 1 UUUUAUAGCUAAGUCUAUAAC-----UUCAAGCUAGGAAACAGCUCAAUGGAUCUUAGAAACAACUUU-CUUCCAAUAAAAUACGAUUGGAAUG--------------GCCUUAAA .((((..((((..((((....-----((((((((.(....)))))...))))...)))).........-.(((((((........)))))))))--------------))..)))) ( -19.70) >DroYak_CAF1 3220 116 - 1 UCUCAUAAGUAAAUCUACAAUCAAAGAUGAAGCUGGGAAACAGCUCAAUGGAUAUUAGAAACAACUUCAAUUCCAAUAAAAUACGAUUGGAAUGGAAGUGGUAGUUGGGCCUUAAA .((((........((((..(((...(.(((.((((.....))))))).).)))..)))).((.(((((.((((((((........)))))))).))))).))...))))....... ( -27.90) >consensus UUUCAAAACUAAAUCUAUAACCAAACAUCAAGCUAGGAAAUAGCUUAAUGGAUCUAAGAAACAAAUCCGAUUCCAAUAAAAUACGAUUGGAAUGGAAGUGGAAGUUAGGCCUUAAA .............................((((((.....))))))...((.(((((....((..((((.(((((((........)))))))))))..))....)))))))..... (-14.28 = -16.24 + 1.96)

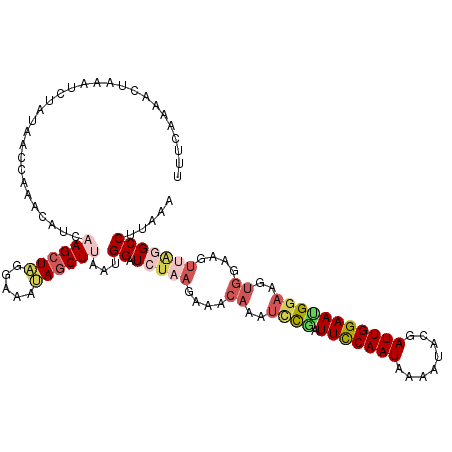
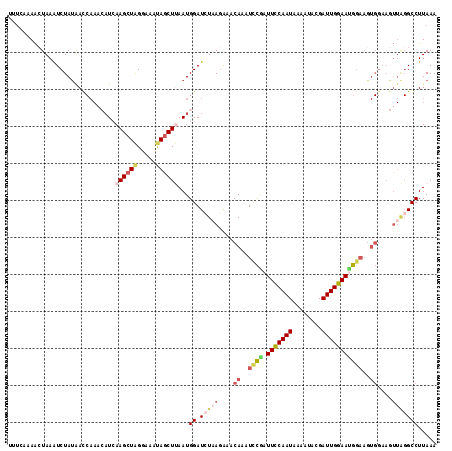
| Location | 15,751,564 – 15,751,660 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 70.60 |
| Mean single sequence MFE | -19.57 |
| Consensus MFE | -13.79 |
| Energy contribution | -16.23 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
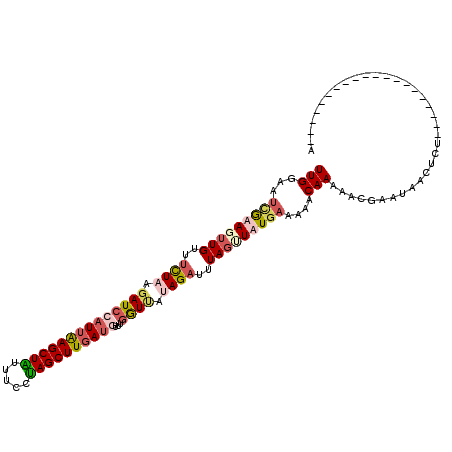
>2L_DroMel_CAF1 15751564 96 + 22407834 UUGAAAUCGGAUUUGUUCUUUAGAUCCAUUAAGCUAUUUUCUAGCUUGAUGUUUGGUUAUAGAUUUAGUUUUGAAACACAAACACGAAUAACUCU--------------------G .......((((.((((((.........(((((((((.....)))))))))((((((((.((((......)))).))).)))))..)))))).)))--------------------) ( -23.50) >DroSim_CAF1 3269 96 + 1 UUGGAAUCGGAUUUGUUUCUUAGAUCCAUUAAGCUAUUUCCUAGCUUGAUGUUUGGUCAUAGAUUUAGUUUUGAAACUCAAAAACGCAUAACUCU--------------------G ...((((((((((((.....)))))))(((((((((.....)))))))))...........))))).(((((........)))))..........--------------------. ( -21.80) >DroEre_CAF1 3323 110 + 1 UUGGAAG-AAAGUUGUUUCUAAGAUCCAUUGAGCUGUUUCCUAGCUUGAA-----GUUAUAGACUUAGCUAUAAAAAACAAAAACGAAUAGAUUUCUGAUAAUAGAUAAUAUAAAA (((..((-(((.((((((..........(..(((((.....)))))..).-----.((((((......))))))...........)))))).)))))..))).............. ( -16.50) >DroYak_CAF1 3260 96 + 1 UUGGAAUUGAAGUUGUUUCUAAUAUCCAUUGAGCUGUUUCCCAGCUUCAUCUUUGAUUGUAGAUUUACUUAUGAGAAACAAAAACGAAUAACUUU--------------------A ......(((...((((((((.(((......((((((.....)))))).((((........)))).....))).))))))))...)))........--------------------. ( -16.50) >consensus UUGGAAUCGAAGUUGUUUCUAAGAUCCAUUAAGCUAUUUCCUAGCUUGAUGUUUGGUUAUAGAUUUAGUUAUGAAAAACAAAAACGAAUAACUCU____________________A (((...(((((((((..((((.((((((((((((((.....))))))))))...)))).))))..)))))))))....)))................................... (-13.79 = -16.23 + 2.44)

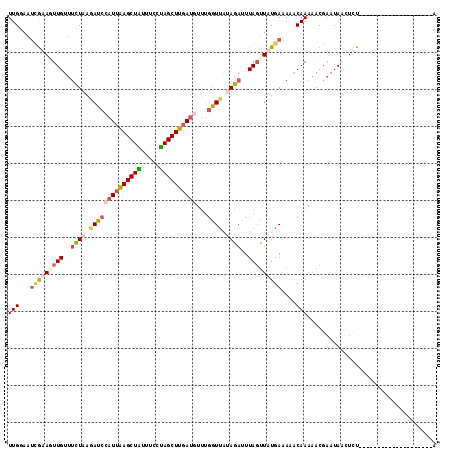
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:46 2006