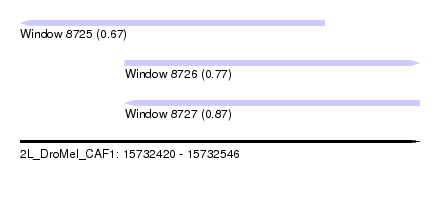
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,732,420 – 15,732,546 |
| Length | 126 |
| Max. P | 0.872020 |

| Location | 15,732,420 – 15,732,516 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 82.85 |
| Mean single sequence MFE | -22.24 |
| Consensus MFE | -15.57 |
| Energy contribution | -16.58 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15732420 96 - 22407834 UCUAGAUGGGUAUCUUUAUCUUUGCAAGAUUAUGAAUAUCUAUAAGUUUUCAUAUAUUAAAUCUUAGGCAUGUUAAAAGGACUGUUUUAGAAC-UCA------ ((((((..(((.(((((.....(((.((((((((((.((......)).))))))......))))...))).....))))))))..))))))..-...------ ( -15.90) >DroSec_CAF1 51931 103 - 1 UCUAAAUGGGUAUCUUUAUCUAUGCAAGAGUUUGAAUAUCUAGAAGUUUUCUUAAAUUAAAUGUAAGGCAUGUUAGGAGGACCUUUCUGGAACAUCAGAUUUU .....(((((((....)))))))...(((((((((...(((((((((((((((((.....(((.....))).))))))))))..)))))))...))))))))) ( -27.20) >DroSim_CAF1 44650 103 - 1 UCUAAAUAGGUAUCUUUAUCUAUGCCAGAGUUUGAAUAUCUAGAAGUUUUCUUAUAUUAAAUAUAAGGCAUGUUAAGAGGACUGUUCUGGAGCAUCAACUUUU .....(((((((....)))))))...((((((.((...((((((((((((((((...................))))))))))..))))))...)))))))). ( -23.61) >consensus UCUAAAUGGGUAUCUUUAUCUAUGCAAGAGUUUGAAUAUCUAGAAGUUUUCUUAUAUUAAAUAUAAGGCAUGUUAAGAGGACUGUUCUGGAACAUCA__UUUU (((..(((((((....)))))))...)))..((((...((((((((((((((((...................)))))))))..)))))))...))))..... (-15.57 = -16.58 + 1.01)


| Location | 15,732,453 – 15,732,546 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 71.74 |
| Mean single sequence MFE | -15.00 |
| Consensus MFE | -6.30 |
| Energy contribution | -6.14 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15732453 93 + 22407834 GAUUUAAUAU-----AUGAAAACUUAUAG---AUAUUCAUAAUCUUGCAAAGAUAAAGAUACCCAUCUAGAGGGUACCAAACUAUCAAACAAGAAAACAAU .........(-----(((((.........---...)))))).(((((....((((..(.(((((.......))))).)....))))...)))))....... ( -13.10) >DroSec_CAF1 51971 93 + 1 CAUUUAAUUU-----AAGAAAACUUCUAG---AUAUUCAAACUCUUGCAUAGAUAAAGAUACCCAUUUAGAGGGUAGCAAACUAUCAAACAAGAAAACAAU ......((((-----(.((.....)))))---))........(((((....((((....(((((.......)))))......))))...)))))....... ( -12.10) >DroSim_CAF1 44690 93 + 1 UAUUUAAUAU-----AAGAAAACUUCUAG---AUAUUCAAACUCUGGCAUAGAUAAAGAUACCUAUUUAGAGGGUACCAAACUAUCAAACAAGAAAACAAU ..........-----.......(((((((---(((.((....((((...))))....))....))))))))))............................ ( -9.30) >DroEre_CAF1 50953 97 + 1 ----CACGAGCAAAACUAAAAAGUUGUAGUUUGUAUUUAUACUCUGGUAUAAAUGAAGAUACCCUUCCAUAGGGUACCAAACUAACCAGCGGGCAAACCAU ----..................((((((((((((((((((((....)))))))))....((((((.....)))))).)))))))..))))((.....)).. ( -25.90) >DroYak_CAF1 56699 89 + 1 ----UAAGACCAGAACUGAAAAGCCG--------AUCUAUACUCUGGUAUAAUUCAAGAUACCCUUCCAGAGGGUACCAAACCAACAAACAAGAAAACCAU ----....((((((...((.......--------.)).....)))))).........(.(((((((...))))))).)....................... ( -14.60) >consensus _AUUUAAUAU_____AUGAAAACUUCUAG___AUAUUCAUACUCUGGCAUAGAUAAAGAUACCCAUCUAGAGGGUACCAAACUAUCAAACAAGAAAACAAU .........................................................(.(((((.......))))).)....................... ( -6.30 = -6.14 + -0.16)

| Location | 15,732,453 – 15,732,546 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 71.74 |
| Mean single sequence MFE | -20.16 |
| Consensus MFE | -10.72 |
| Energy contribution | -11.92 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.872020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15732453 93 - 22407834 AUUGUUUUCUUGUUUGAUAGUUUGGUACCCUCUAGAUGGGUAUCUUUAUCUUUGCAAGAUUAUGAAUAU---CUAUAAGUUUUCAU-----AUAUUAAAUC .......((((((..(((((...(((((((.......))))))).)))))...)))))).((((((.((---......)).)))))-----)......... ( -24.00) >DroSec_CAF1 51971 93 - 1 AUUGUUUUCUUGUUUGAUAGUUUGCUACCCUCUAAAUGGGUAUCUUUAUCUAUGCAAGAGUUUGAAUAU---CUAGAAGUUUUCUU-----AAAUUAAAUG .(((..(((((((..(((((...(.(((((.......))))).).)))))...)))))))..)))....---..............-----.......... ( -18.50) >DroSim_CAF1 44690 93 - 1 AUUGUUUUCUUGUUUGAUAGUUUGGUACCCUCUAAAUAGGUAUCUUUAUCUAUGCCAGAGUUUGAAUAU---CUAGAAGUUUUCUU-----AUAUUAAAUA ..........((((((((((((..(....((((..(((((((....)))))))...)))).)..)))..---....(((....)))-----.))))))))) ( -14.70) >DroEre_CAF1 50953 97 - 1 AUGGUUUGCCCGCUGGUUAGUUUGGUACCCUAUGGAAGGGUAUCUUCAUUUAUACCAGAGUAUAAAUACAAACUACAACUUUUUAGUUUUGCUCGUG---- ..((....))(((.(((((((((((((((((.....)))))))....((((((((....)))))))).))))))).((((....))))..))).)))---- ( -24.00) >DroYak_CAF1 56699 89 - 1 AUGGUUUUCUUGUUUGUUGGUUUGGUACCCUCUGGAAGGGUAUCUUGAAUUAUACCAGAGUAUAGAU--------CGGCUUUUCAGUUCUGGUCUUA---- ......((((.((....((((((((((((((.....))))))))..)))))).)).))))...((((--------(((..(....)..)))))))..---- ( -19.60) >consensus AUUGUUUUCUUGUUUGAUAGUUUGGUACCCUCUAGAUGGGUAUCUUUAUCUAUGCCAGAGUAUGAAUAU___CUACAAGUUUUCAU_____AUAUUAAAU_ ......((((.((..(((((...(((((((.......))))))).)))))...)).))))......................................... (-10.72 = -11.92 + 1.20)

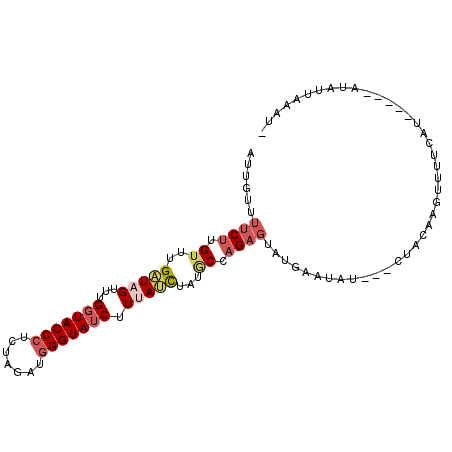
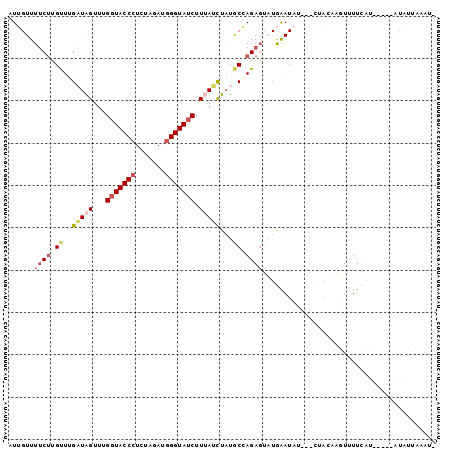
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:40 2006