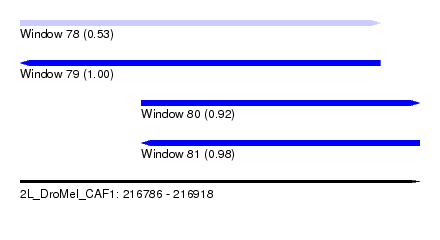
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 216,786 – 216,918 |
| Length | 132 |
| Max. P | 0.996216 |

| Location | 216,786 – 216,905 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.09 |
| Mean single sequence MFE | -49.70 |
| Consensus MFE | -25.07 |
| Energy contribution | -25.27 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.50 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 216786 119 + 22407834 GACAUUCCGGGAAGCAGAGACGAUAGGGAGGUGGCAUUUCCACCUGCUGCAGCUGCCGCAACGGCCGCUGCCACAGCAGCAUUGAUGCUUUUAGCAUCCAAGCCAGGUAAACU-GUUGGA .....(((((...((..........((.((((((.....)))))).))(((((.((((...)))).)))))....)).((...(((((.....)))))...))(((.....))-)))))) ( -41.10) >DroVir_CAF1 11967 119 + 1 GACAUGCCCGGCAGCAAUGCCGACAAGGAUGUGGCAUUGCCGCCGCUUGCCGCAGCGGCGACCGCAGCCGCGACAGCUGCAUUGAUGCUAUUCGCAUCCAGGCCCGGCAGCGUGGUGG-G ........((((.((((((((.(((....))))))))))).))))((..((((.(((((.......)))))....(((((.(((((((.....)))).))).....)))))))))..)-) ( -58.60) >DroGri_CAF1 10362 119 + 1 GACAUGCCCGGCAGCAGGGCAGAGAGCGAGGUGGCAUUGCCACCACUAGCGGCAGCCGCAACAGCAGCUGCCACUGCUGCAUUUAUGCUAUUCGCAUCCAGGCCAGGCAGCGUUGUUG-U (((.((((.(((((((((((((...((..((((((...))))))....((((...))))....))..))))).)))))......((((.....))))....))).))))..)))....-. ( -52.60) >DroWil_CAF1 10817 117 + 1 GACAUGCCCGGCAGGAGGGAUGAUAGCGAUGUGGCAUUGCCACCAGCUGCAGCAGCUGCCACAGCAGCAGCAACUGCCGCAUUGAUGCUAUUGGCAUCGAGUCCUGGUAA--U-GUUGGU ((((((((((((((......((.((((...(((((...)))))..))))))((.(((((....))))).))..))))))..(((((((.....))))))).....))).)--)-)))... ( -48.40) >DroMoj_CAF1 9755 119 + 1 GACAUGCCCGGCAGCAGAGCUGACAGAGAUGUGGCAUUGCCGCCACUGGCUGCCGCUGCGACAGCAGCUGCCACAGCCGCAUUGAUGCUGUUCGCAUCCAAACCAGGCAGCGUUGUUG-G .......((((((((.((((.(.((..((((((((...((((....)))).((.(((((....))))).))....))))))))..))).))))((..((......))..)))))))))-) ( -49.50) >DroAna_CAF1 12042 116 + 1 GACAUGCCAGGCAGCAGGCUGGACAGCGAGGUGGCAUUACCACCUGCCGCAGCUGCCGCCACAGCAGCCGCCACCGCAGCAUUGAUGUUGUUCGCAUCCAGUAGCGGUAGAGU----GGA ....(((..(((.((((.(((....((.((((((.....))))))))..))))))).)))...))).((((((((((.((...(((((.....)))))..)).))))).).))----)). ( -48.00) >consensus GACAUGCCCGGCAGCAGGGCCGACAGCGAGGUGGCAUUGCCACCACCUGCAGCAGCCGCAACAGCAGCUGCCACAGCAGCAUUGAUGCUAUUCGCAUCCAGGCCAGGCAGCGU_GUUG_A ....((((..((.((...............((((.....)))).....((.((.((((...)))).)).))....)).))...(((((.....))))).......))))........... (-25.07 = -25.27 + 0.20)

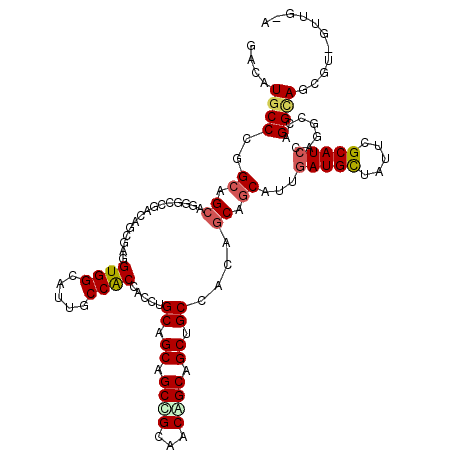
| Location | 216,786 – 216,905 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.09 |
| Mean single sequence MFE | -55.53 |
| Consensus MFE | -34.13 |
| Energy contribution | -34.24 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 216786 119 - 22407834 UCCAAC-AGUUUACCUGGCUUGGAUGCUAAAAGCAUCAAUGCUGCUGUGGCAGCGGCCGUUGCGGCAGCUGCAGCAGGUGGAAAUGCCACCUCCCUAUCGUCUCUGCUUCCCGGAAUGUC (((.((-(((......(((...(((((.....)))))...))))))))(((((.(((.(((((((...)))))))((((((.....)))))).......))).)))))....)))..... ( -44.80) >DroVir_CAF1 11967 119 - 1 C-CCACCACGCUGCCGGGCCUGGAUGCGAAUAGCAUCAAUGCAGCUGUCGCGGCUGCGGUCGCCGCUGCGGCAAGCGGCGGCAAUGCCACAUCCUUGUCGGCAUUGCUGCCGGGCAUGUC (-((.....((((((.((((..(((((.....)))))...(((((((...)))))))))))((....))))).)))(((((((((((((((....))).)))))))))))))))...... ( -65.20) >DroGri_CAF1 10362 119 - 1 A-CAACAACGCUGCCUGGCCUGGAUGCGAAUAGCAUAAAUGCAGCAGUGGCAGCUGCUGUUGCGGCUGCCGCUAGUGGUGGCAAUGCCACCUCGCUCUCUGCCCUGCUGCCGGGCAUGUC .-.....(((.((((((((..((((((.....))))....((((.((((((((((((....)))))))))))).(.((((((...)))))).).....))))))....))))))))))). ( -60.90) >DroWil_CAF1 10817 117 - 1 ACCAAC-A--UUACCAGGACUCGAUGCCAAUAGCAUCAAUGCGGCAGUUGCUGCUGCUGUGGCAGCUGCUGCAGCUGGUGGCAAUGCCACAUCGCUAUCAUCCCUCCUGCCGGGCAUGUC ....((-(--(..((((((...((((...(((((..((.((((((((((((..(....)..))))))))))))..))(((((...)))))...)))))))))..))))...))..)))). ( -50.20) >DroMoj_CAF1 9755 119 - 1 C-CAACAACGCUGCCUGGUUUGGAUGCGAACAGCAUCAAUGCGGCUGUGGCAGCUGCUGUCGCAGCGGCAGCCAGUGGCGGCAAUGCCACAUCUCUGUCAGCUCUGCUGCCGGGCAUGUC .-.....(((.((((((((...(((((.....)))))...(((((((((((.(((((((...))))))).))))((((((....))))))........)))))..)).))))))))))). ( -55.40) >DroAna_CAF1 12042 116 - 1 UCC----ACUCUACCGCUACUGGAUGCGAACAACAUCAAUGCUGCGGUGGCGGCUGCUGUGGCGGCAGCUGCGGCAGGUGGUAAUGCCACCUCGCUGUCCAGCCUGCUGCCUGGCAUGUC ...----...(((((((..(..((((.......))))...)..))))))).((((((((.(((((((((((((((((((((.....)))))).))))..))).))))))))))))).))) ( -56.70) >consensus A_CAAC_ACGCUACCUGGCCUGGAUGCGAAUAGCAUCAAUGCAGCUGUGGCAGCUGCUGUCGCGGCUGCUGCAACUGGUGGCAAUGCCACAUCGCUGUCAGCCCUGCUGCCGGGCAUGUC .........((((((.......(((((.....)))))...((.(((((.(((((....))))).))))).))....)))))).(((((........................)))))... (-34.13 = -34.24 + 0.11)
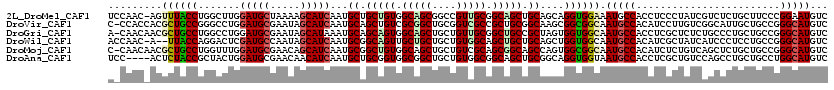
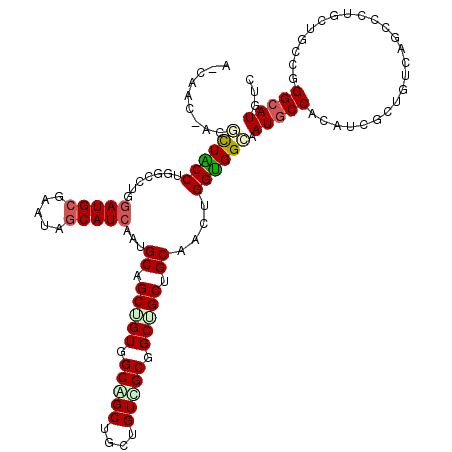
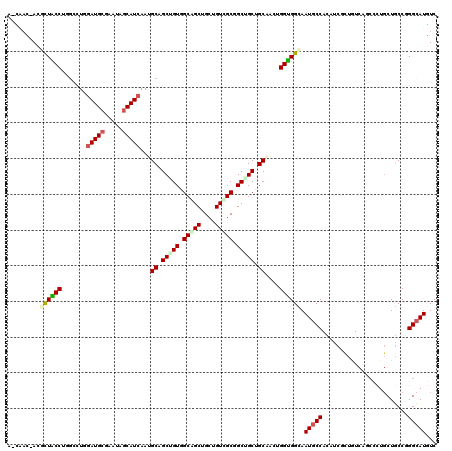
| Location | 216,826 – 216,918 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 76.56 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -20.67 |
| Energy contribution | -21.58 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 216826 92 + 22407834 CACCUGCUGCAGCUGCCGCAACGGCCGCUGCCACAGCAGCAUUGAUGCUUUUAGCAUCCAAGCCAGGUAAACU-GUUGGAAGAA---GAGGGCUUU ...((((((((((.((((...)))).)))))...))))).......((((((..(.(((((..(((.....))-)))))).)..---))))))... ( -34.40) >DroSim_CAF1 9116 92 + 1 CACCUGCCGCAGCUGCCGCAACGGCCGCUGCCACAGCAGCAUUAAUGCUUUUGGCAUCCAAGCCAGGCAAACU-GUUGGAAGAA---GAGGGCUUU .....((((((((.((((...)))).)))))..((((((......((((..((((......))))))))..))-))))......---...)))... ( -35.00) >DroEre_CAF1 9407 92 + 1 CACCUGCUGCAGCUGCCGCAACGGCUGCUGCCACCGCAGCAUUGAUGCUUUUGGCAUCCAAUCCAGGUAAGCU-GUUGGAAGAA---GACGGCUUU .(((((..(((((.((((...)))).)))))............(((((.....))))).....)))))(((((-(((.......---)))))))). ( -35.20) >DroYak_CAF1 11169 92 + 1 CACCUGCUGCAGCUGCCGCAACGGCCGCUGCCACCGCAGCAUUGAUGCUUUUGGCAUCCAAGCCAGGUAAGCU-GUUGGAAGAG---GAGGGUUUU ..(((.(((((((.((((...)))).)))))..(((((((.....((((..((((......)))))))).)))-).)))...))---.)))..... ( -35.70) >DroMoj_CAF1 9795 86 + 1 CGCCACUGGCUGCCGCUGCGACAGCAGCUGCCACAGCCGCAUUGAUGCUGUUCGCAUCCAAACCAGGCAGCGUUGUUG-GCGCUGUU--------- ((((((..(((((((((((....)))))(((.(((((.........)))))..))).........))))))...).))-))).....--------- ( -36.30) >DroAna_CAF1 12082 92 + 1 CACCUGCCGCAGCUGCCGCCACAGCAGCCGCCACCGCAGCAUUGAUGUUGUUCGCAUCCAGUAGCGGUAGAGU----GGAAGCACUCACGGGCUUG .....(((((.(((((.......))))).)).(((((.((...(((((.....)))))..)).))))).((((----(....)))))...)))... ( -36.30) >consensus CACCUGCUGCAGCUGCCGCAACGGCCGCUGCCACAGCAGCAUUGAUGCUUUUGGCAUCCAAGCCAGGUAAACU_GUUGGAAGAA___GAGGGCUUU .(((((..(((((.((((...)))).)))))............(((((.....))))).....)))))............................ (-20.67 = -21.58 + 0.92)

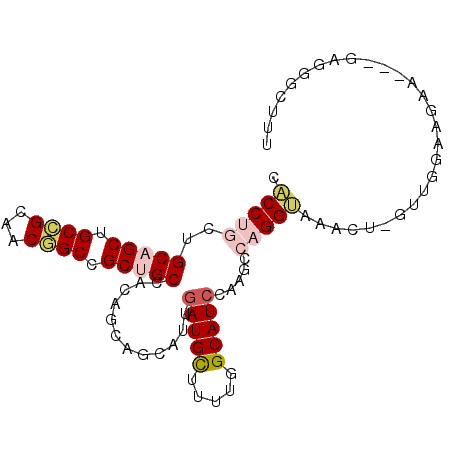
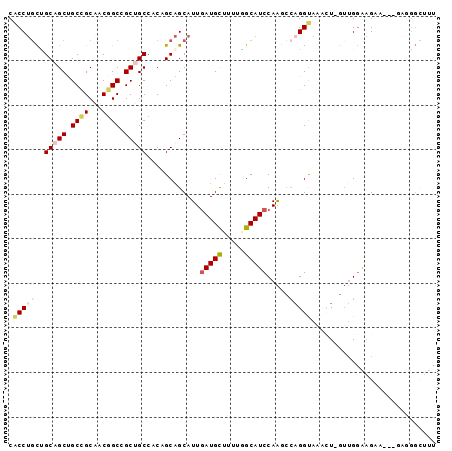
| Location | 216,826 – 216,918 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 76.56 |
| Mean single sequence MFE | -37.18 |
| Consensus MFE | -27.66 |
| Energy contribution | -27.42 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 216826 92 - 22407834 AAAGCCCUC---UUCUUCCAAC-AGUUUACCUGGCUUGGAUGCUAAAAGCAUCAAUGCUGCUGUGGCAGCGGCCGUUGCGGCAGCUGCAGCAGGUG .........---..........-....((((((.....(((((.....)))))..(((.(((((.(((((....))))).))))).))).)))))) ( -35.90) >DroSim_CAF1 9116 92 - 1 AAAGCCCUC---UUCUUCCAAC-AGUUUGCCUGGCUUGGAUGCCAAAAGCAUUAAUGCUGCUGUGGCAGCGGCCGUUGCGGCAGCUGCGGCAGGUG ...(((...---....((((.(-((.....)))...))))((((...((((....))))(((((.(((((....))))).)))))...))))))). ( -35.00) >DroEre_CAF1 9407 92 - 1 AAAGCCGUC---UUCUUCCAAC-AGCUUACCUGGAUUGGAUGCCAAAAGCAUCAAUGCUGCGGUGGCAGCAGCCGUUGCGGCAGCUGCAGCAGGUG .((((.((.---........))-.))))(((((.....(((((.....)))))..((((((..((((....))))..)))))).......))))). ( -34.20) >DroYak_CAF1 11169 92 - 1 AAAACCCUC---CUCUUCCAAC-AGCUUACCUGGCUUGGAUGCCAAAAGCAUCAAUGCUGCGGUGGCAGCGGCCGUUGCGGCAGCUGCAGCAGGUG ...(((...---..........-.((((...((((......)))).)))).....(((((((((.(((((....)))))....)))))))))))). ( -33.80) >DroMoj_CAF1 9795 86 - 1 ---------AACAGCGC-CAACAACGCUGCCUGGUUUGGAUGCGAACAGCAUCAAUGCGGCUGUGGCAGCUGCUGUCGCAGCGGCAGCCAGUGGCG ---------.....(((-((.....((((((..((...(((((.....)))))...)).((((((((((...))))))))))))))))...))))) ( -44.30) >DroAna_CAF1 12082 92 - 1 CAAGCCCGUGAGUGCUUCC----ACUCUACCGCUACUGGAUGCGAACAACAUCAAUGCUGCGGUGGCGGCUGCUGUGGCGGCAGCUGCGGCAGGUG ...(((...(((((....)----))))((((((..(..((((.......))))...)..))))))((((((((((...)))))))))))))..... ( -39.90) >consensus AAAGCCCUC___UUCUUCCAAC_AGUUUACCUGGCUUGGAUGCCAAAAGCAUCAAUGCUGCGGUGGCAGCGGCCGUUGCGGCAGCUGCAGCAGGUG ............................(((.......(((((.....)))))..(((((((((.((.(((.....))).)).)))))))))))). (-27.66 = -27.42 + -0.25)

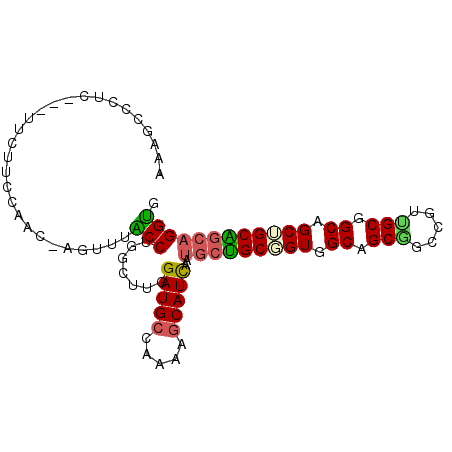
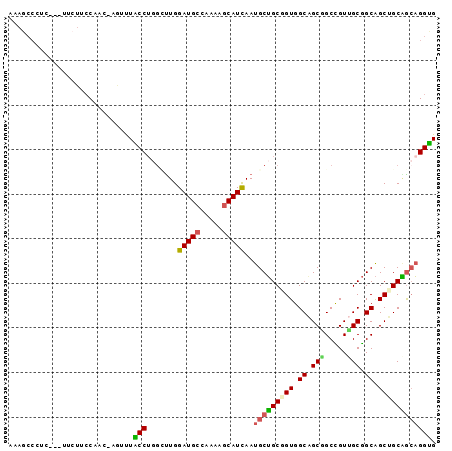
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:28 2006