| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
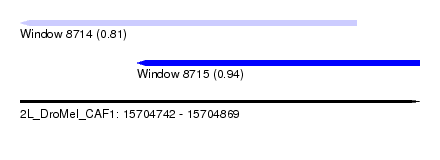
| Location | 15,704,742 – 15,704,869 |
| Length | 127 |
| Max. P | 0.939671 |

| Location | 15,704,742 – 15,704,849 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.70 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -19.83 |
| Energy contribution | -20.42 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
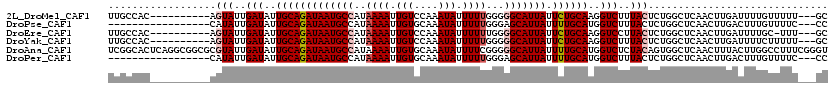
>2L_DroMel_CAF1 15704742 107 - 22407834 UUGCCAC----------AGUAUUGAUAUUGCAGAUAAUGCCAUAAAAUUGUCCAAAUAUUUUUGGGGGCAUUAUUCUGCAAGGUCUUUACUCUGGCUCAACUUGAUUUUGUUUUU---GC ..((((.----------((((..(((.((((((((((((((.........((((((....))))))))))))).))))))).)))..)))).))))...................---.. ( -37.20) >DroPse_CAF1 31994 100 - 1 -----------------CAUAUUGAUAUUGCAGAUAAUGCCAUAAAAUUGUGCAAAUAUUUUUGGGAGCAUUAUUUUGCAUGGUCUUUACUCUGGCUCAACUUGACUUUGUUUUC---CC -----------------((..((((...(((((((((((((.(((((.(((....))).))))).).)))))).))))))..(((........)))))))..))...........---.. ( -19.40) >DroEre_CAF1 32657 106 - 1 UUGCCAC----------AGUAUUGAUAUUGCAGAUAAUGCCAUAAAAUUGUCCAAAUAUUUUUUGGGGCAUUAUUCUGCAAGGUCCUUACUCUGGCUCAACUUGAUUUUGC-UUU---GC ..((((.----------((((..(((.(((((((((((((((((....)))(((((.....)))))))))))).))))))).)))..)))).))))...............-...---.. ( -35.60) >DroYak_CAF1 35674 107 - 1 UUGCCAC----------AGUAUUGAUAUUGCAGAUAAUGCCAUAAAAUUGUCCAAAUAUUUUUGGGGGCAUUAUUCUGCAAGGUCUUUACUCUGGCUCAACUUGAUUUUCUUUUU---GC ..((((.----------((((..(((.((((((((((((((.........((((((....))))))))))))).))))))).)))..)))).))))...................---.. ( -37.20) >DroAna_CAF1 24252 120 - 1 UCGGCACUCAGGCGGCGCGUAUUGAUAUUGCAGAUAAUGCCAUAAAAUUGUGCAAAUAUUUUCGGGGGCAUUAUUUUGCAUGGUCUCUACAGUGGCUCAACUUUACUUGGCCUUUCGGGU .....(((((((((.((((((..(((..(((((((((((((...(((.(((....))).)))....))))))).))))))..)))..))).))).).(((......)))))))...)))) ( -32.50) >DroPer_CAF1 29735 100 - 1 -----------------CAUAUUGAUAUUGCAGAUAAUGCCAUAAAAUUGUGCAAAUAUUUUUGGGAGCAUUAUUUUGCAUGGUCUUUACUCUGGCUCAACUUGACUUUGUUUUC---CC -----------------((..((((...(((((((((((((.(((((.(((....))).))))).).)))))).))))))..(((........)))))))..))...........---.. ( -19.40) >consensus UUGCCAC__________AGUAUUGAUAUUGCAGAUAAUGCCAUAAAAUUGUCCAAAUAUUUUUGGGGGCAUUAUUCUGCAAGGUCUUUACUCUGGCUCAACUUGACUUUGUUUUU___GC ..................(((..(((..(((((((((((((..((((.(((....))).))))...))))))).))))))..)))..))).............................. (-19.83 = -20.42 + 0.58)


| Location | 15,704,779 – 15,704,869 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -18.97 |
| Consensus MFE | -15.57 |
| Energy contribution | -15.68 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15704779 90 - 22407834 CUUCACCUACCUCCCUCUCUUUGCCACAGUAUUGAUAUUGCAGAUAAUGCCAUAAAAUUGUCCAAAUAUUUUUGGGGGCAUUAUUCUGCA .................((..(((....)))..))...(((((((((((((.........((((((....))))))))))))).)))))) ( -21.50) >DroPse_CAF1 32031 72 - 1 CCU------CCUCCC------------CAUAUUGAUAUUGCAGAUAAUGCCAUAAAAUUGUGCAAAUAUUUUUGGGAGCAUUAUUUUGCA ...------....((------------((....(((((((((..((((........))))))).))))))..)))).(((......))). ( -14.70) >DroSim_CAF1 25539 90 - 1 CUUCACCUACCUCCCUCUCUUUGCCACAGUAUUGAUAUUGCAGAUAAUGCCAUAAAAUUGUCCAAAUAUUUUUGGGGGCAUUAUUCUGCA .................((..(((....)))..))...(((((((((((((.........((((((....))))))))))))).)))))) ( -21.50) >DroEre_CAF1 32693 90 - 1 CUUCACCCACCUCCCUCUCCUUGCCACAGUAUUGAUAUUGCAGAUAAUGCCAUAAAAUUGUCCAAAUAUUUUUUGGGGCAUUAUUCUGCA .................((..(((....)))..))...((((((((((((((((....)))(((((.....)))))))))))).)))))) ( -19.90) >DroYak_CAF1 35711 90 - 1 CUUUACCUACCUUCCCCUCUUUGCCACAGUAUUGAUAUUGCAGAUAAUGCCAUAAAAUUGUCCAAAUAUUUUUGGGGGCAUUAUUCUGCA .................((..(((....)))..))...(((((((((((((.........((((((....))))))))))))).)))))) ( -21.50) >DroPer_CAF1 29772 72 - 1 CCU------CCUCCC------------CAUAUUGAUAUUGCAGAUAAUGCCAUAAAAUUGUGCAAAUAUUUUUGGGAGCAUUAUUUUGCA ...------....((------------((....(((((((((..((((........))))))).))))))..)))).(((......))). ( -14.70) >consensus CUUCACCUACCUCCCUCUCUUUGCCACAGUAUUGAUAUUGCAGAUAAUGCCAUAAAAUUGUCCAAAUAUUUUUGGGGGCAUUAUUCUGCA ......................................(((((((((((((..((((.(((....))).))))...))))))).)))))) (-15.57 = -15.68 + 0.11)

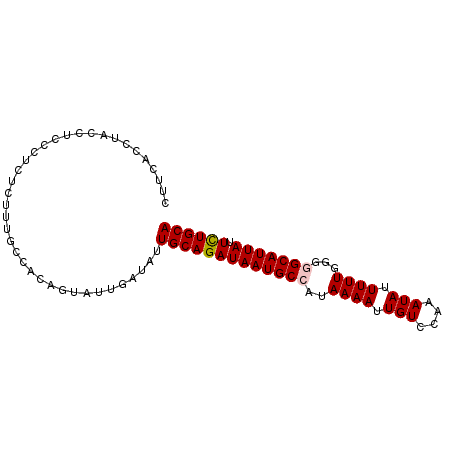
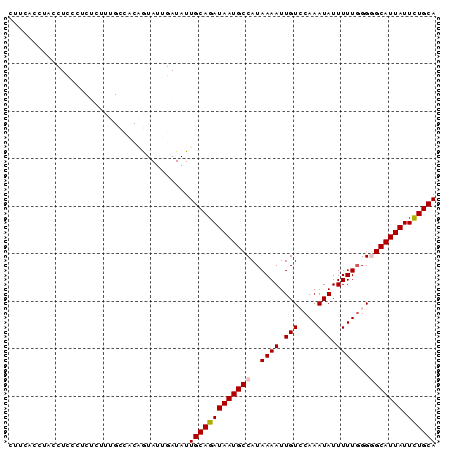
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:28 2006