| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,702,686 – 15,702,780 |
| Length | 94 |
| Max. P | 0.808326 |

| Location | 15,702,686 – 15,702,780 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.48 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -9.56 |
| Energy contribution | -10.62 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
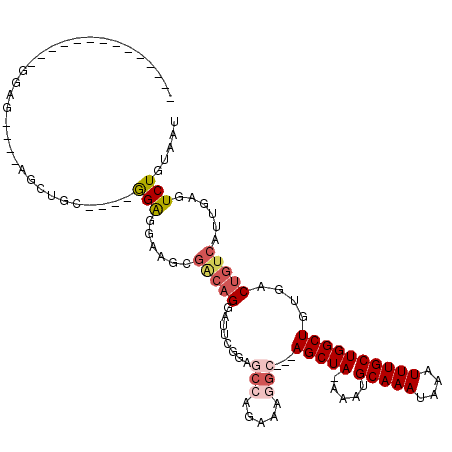
>2L_DroMel_CAF1 15702686 94 - 22407834 --------------GGAG----AGCUGC----GGAGGAAGCGACAGGAUUCGGAGCCAGAAAGGC---AGCUA-AAAUGCAAAUAAAUUUGCUGGCUGUGACUGUCAUUGAGUCUGUAAU --------------....----...(((----(((...((.(((((...(((.(((((((((.((---(....-...))).......))).)))))).)))))))).))...)))))).. ( -26.60) >DroPse_CAF1 29737 103 - 1 AUCCACAGUCAGAAUAAGAGUCAGGGGC----AGAGGCAGGGGCAG-AGU------CAG--AGGC---AGCUA-AAAUGCAAAUAAAUUUGCUGGCUGUGACUGUCAUUGAGUCUGUAAU ..((..(.((.......)).)...))((----(((..(((.((((.-.((------((.--((.(---(((..-((((........)))))))).)).)))))))).)))..)))))... ( -29.60) >DroSec_CAF1 31072 94 - 1 --------------GGCG----AGCUGC----GGAGGAAGCGACAGGAUUCGCAGCCAGAAAGGC---AGCUA-AAAUGCAAAUAAAUUUGCUGGCUGUGACUGUCAUUGAGUCUGUAAU --------------(((.----.(((..----......)))(((((...(((((((((((((.((---(....-...))).......))).))))))))))))))).....)))...... ( -31.50) >DroYak_CAF1 33603 94 - 1 --------------GGAG----UGCUGC----GGAGGAAGCGACAGGAUUCGGAGCCAGAAAAGC---AGCUA-AAAUGCAAAUAAAUUUGCUGGCUGCGACUGUCAUUGAGUCUGUAAU --------------....----((((..----......))))((((.((((((.(.(((....((---(((((-....(((((....))))))))))))..))).).))))))))))... ( -27.90) >DroMoj_CAF1 31821 78 - 1 --------------------------AG----GGAGGCAGCAGCAG---------C-CC--AGGCUGAAGCUAAAAAUGCAAAUAAAUUUGCUGGCUGUGACUGUCAUUGAGUCUGUAAU --------------------------..----...(((((..((((---------(-(.--.(((....)))......(((((....))))).))))))..))))).............. ( -24.20) >DroAna_CAF1 22462 98 - 1 --------------GGAG----AGAAGCAGUUGGGGGAUGGGUGCGAAGUCGGUGCCAGAAAGGC---AGCUA-AAAUGCAAAUAAAUUUGCUGGCUGUGACUGUCAUUGAGUCUGUAAU --------------.((.----((..(((((..(.((((...((((.....(.((((.....)))---).)..-...)))).....)))).)..)))))..)).)).............. ( -29.00) >consensus ______________GGAG____AGCUGC____GGAGGAAGCGACAGGAUUCGGAGCCAGAAAGGC___AGCUA_AAAUGCAAAUAAAUUUGCUGGCUGUGACUGUCAUUGAGUCUGUAAU ................................(((......(((((........(((.....)))...(((((.....(((((....))))))))))....)))))......)))..... ( -9.56 = -10.62 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:26 2006