
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,698,700 – 15,698,813 |
| Length | 113 |
| Max. P | 0.771175 |

| Location | 15,698,700 – 15,698,813 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -45.64 |
| Consensus MFE | -25.54 |
| Energy contribution | -27.30 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15698700 113 + 22407834 AGGAUCAAGGACUGUGUGGGUCUGCGGCCC--GAAUGCAUUUUGGACCGCACAAAAGCCAGUGGAGAGUAGCUCCAUGAAGGAGCUG-AGCUGCCCUCAGCCAUUCCGGUGGAUCC---- .(((((..(((((.....)))))((((.((--(((.....))))).))))......(((.((((((.....))))))...(((((((-((.....))))))...)))))).)))))---- ( -45.20) >DroSec_CAF1 27190 113 + 1 AGGAUCAAGGACUGUGUGGGUCUGCGGCCC--GAAUGCAUUUUGGACCGCACAAAAGCCAGUGGAGAGUAGCUCCAUGAAGGAGCUG-AGCUGCCCCCAGCCAUUCCGGUGGAUAC---- ...(((..(((.((..((((..(((((.((--(((.....))))).)))))...........((((..(((((((.....)))))))-..)).))))))..)).)))....)))..---- ( -42.40) >DroEre_CAF1 27433 114 + 1 AGGAUCAAGGACUGUGUGGGUGUGCGGCUC--GAAUGCAUUUUGGACCGCACAAAAGCCAGUGGAGAGUAGCUCCAUAAAGCAGCUUCGGCUGCCCUCAGCCAUUCCGGUGGAUCC---- .(((((..(((.((..(((((((((((..(--(((.....))))..))))))).......((((((.....))))))...(((((....))))).))))..)).)))....)))))---- ( -50.80) >DroYak_CAF1 28662 113 + 1 AGGAUCAACGACUGUGUGGGUCUGCGGCCC--GAAUGCAUUUUGGACCGCACAAAAGCCAGUGGAGAGUAGCUCCAUGAAGGAGCUG-AGCUGGCCUCGGCCAUUCCGUUGGAUCC---- .((((((((.((((.((.....(((((.((--(((.....))))).))))).....))))))((((..(((((((.....)))))))-..)((((....)))).)))))).)))))---- ( -49.30) >DroAna_CAF1 20579 112 + 1 -----GCAGGGCUCUGUGGGUCUGCAGCCCCGUAAUGCAUUUGGGACCCCACAAAAGCCAAUGG--AGUAGCCCCAUGAAGGAGUUG-AGUCUCCCUUAGCCUUUGGGGAGACCACACCG -----...((((((((((((.((((....((((...((.((((((...)).)))).))..))))--.)))).)))))...)))))((-.((((((((........))))))))))..)). ( -40.50) >consensus AGGAUCAAGGACUGUGUGGGUCUGCGGCCC__GAAUGCAUUUUGGACCGCACAAAAGCCAGUGGAGAGUAGCUCCAUGAAGGAGCUG_AGCUGCCCUCAGCCAUUCCGGUGGAUCC____ ..............(((((((((...((........)).....))))).))))....(((.(((((...((((((.....))))))...((((....))))..))))).)))........ (-25.54 = -27.30 + 1.76)

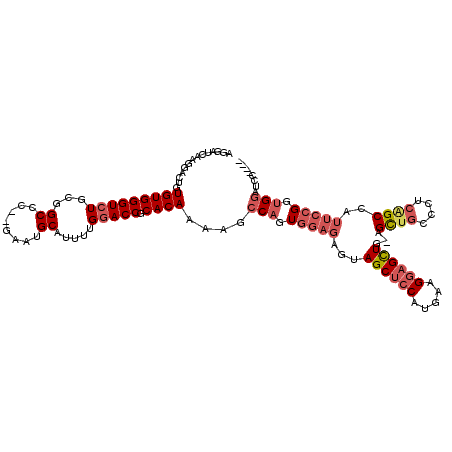
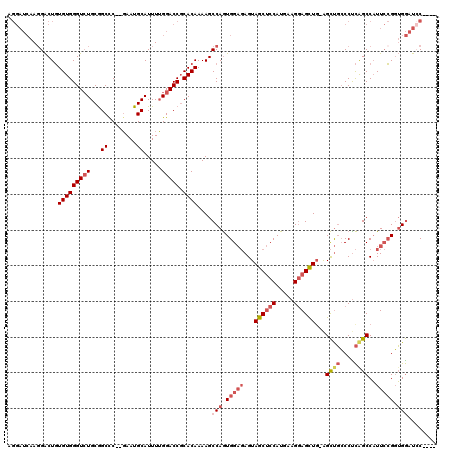
| Location | 15,698,700 – 15,698,813 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -43.10 |
| Consensus MFE | -26.12 |
| Energy contribution | -27.04 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15698700 113 - 22407834 ----GGAUCCACCGGAAUGGCUGAGGGCAGCU-CAGCUCCUUCAUGGAGCUACUCUCCACUGGCUUUUGUGCGGUCCAAAAUGCAUUC--GGGCCGCAGACCCACACAGUCCUUGAUCCU ----(((((....(((..(((((....)))))-.((((((.....))))))....)))((((.....(.((((((((...........--)))))))).)......))))....))))). ( -42.90) >DroSec_CAF1 27190 113 - 1 ----GUAUCCACCGGAAUGGCUGGGGGCAGCU-CAGCUCCUUCAUGGAGCUACUCUCCACUGGCUUUUGUGCGGUCCAAAAUGCAUUC--GGGCCGCAGACCCACACAGUCCUUGAUCCU ----.........(((.((..((((((.((..-.((((((.....))))))...)))).........(.((((((((...........--)))))))).)))))..)).)))........ ( -39.40) >DroEre_CAF1 27433 114 - 1 ----GGAUCCACCGGAAUGGCUGAGGGCAGCCGAAGCUGCUUUAUGGAGCUACUCUCCACUGGCUUUUGUGCGGUCCAAAAUGCAUUC--GAGCCGCACACCCACACAGUCCUUGAUCCU ----(((((....(((.(((((.((((((((....)))))))).(((((.....)))))..)))...(((((((..(...........--)..)))))))......)).)))..))))). ( -41.10) >DroYak_CAF1 28662 113 - 1 ----GGAUCCAACGGAAUGGCCGAGGCCAGCU-CAGCUCCUUCAUGGAGCUACUCUCCACUGGCUUUUGUGCGGUCCAAAAUGCAUUC--GGGCCGCAGACCCACACAGUCGUUGAUCCU ----(((((.((((((.((((....))))...-.((((((.....))))))....)))((((.....(.((((((((...........--)))))))).)......)))).)))))))). ( -47.00) >DroAna_CAF1 20579 112 - 1 CGGUGUGGUCUCCCCAAAGGCUAAGGGAGACU-CAACUCCUUCAUGGGGCUACU--CCAUUGGCUUUUGUGGGGUCCCAAAUGCAUUACGGGGCUGCAGACCCACAGAGCCCUGC----- ..(((.((((((((((((((((((.((((...-...((((.....))))...))--)).)))))))))).)))(((((...........)))))...))))))))..........----- ( -45.10) >consensus ____GGAUCCACCGGAAUGGCUGAGGGCAGCU_CAGCUCCUUCAUGGAGCUACUCUCCACUGGCUUUUGUGCGGUCCAAAAUGCAUUC__GGGCCGCAGACCCACACAGUCCUUGAUCCU ..................(((((.(((.......((((((.....))))))..................((((((((.............))))))))..)))...)))))......... (-26.12 = -27.04 + 0.92)

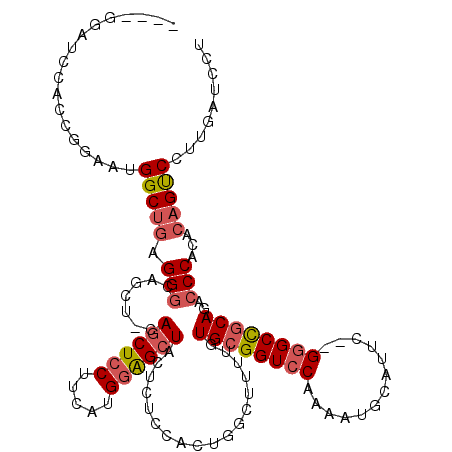
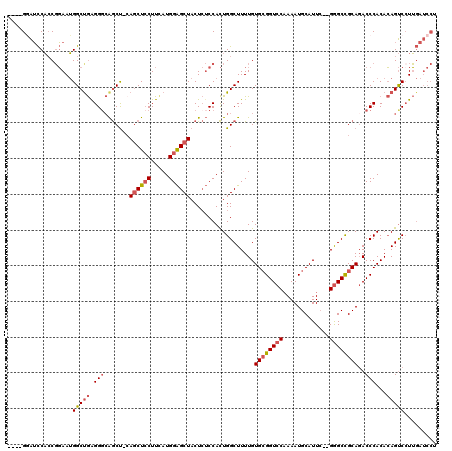
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:24 2006