| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,659,562 – 15,659,652 |
| Length | 90 |
| Max. P | 0.987859 |

| Location | 15,659,562 – 15,659,652 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 70.22 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -19.51 |
| Energy contribution | -18.96 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
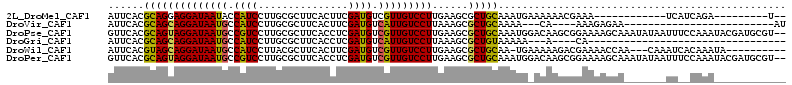
>2L_DroMel_CAF1 15659562 90 + 22407834 AUUCACGCAGGAGGAUAAUACCAUCCUUGCGCUUCACUUCGAUGUCGUUGUCCUUGAAGCGCUGCAAAUGAAAAAACGAAA------------UCAUCAGA---------U-- .((((.(((.((((((......))))))((((((((....(((......)))..)))))))))))...)))).........------------........---------.-- ( -22.90) >DroVir_CAF1 37210 81 + 1 AUUCACGCAGCAGGAUAAUGCCAUCCUUGCGCUUCACUUCGAUGUCAUUGUCCUUAAAGCGCUGCAAAA---CA----AAAGAGAA-------------------------AU ......((((((((((((((.((((...............)))).)))))))))......)))))....---..----........-------------------------.. ( -21.56) >DroPse_CAF1 18302 111 + 1 GUUCACGCAGUAGGAUAAUGCCGUCCUUGCGCUUCACCUCGAUGUCGUUGUCCUUGAAGCGCUGCAAAUGGACAAGCGGAAAAGCAAAUAUAAUUUCCAAAUACGAUGCGU-- ....(((((((.((......))((((..((((((((....(((......)))..)))))))).......))))..))(((((...........)))))........)))))-- ( -29.00) >DroGri_CAF1 20613 73 + 1 AUUCACGCAGCAGGAUAAUGCCAUCCUUGCGCUUCACCUCGAUGUCAUUGUCCUUAAAGCGCUGUAAAAA---A----CA--------------------------------- ......((((((((((((((.((((...............)))).)))))))))......))))).....---.----..--------------------------------- ( -19.16) >DroWil_CAF1 37846 99 + 1 AUUCACGUAGCAGGAUAAUGCCAUCCUUACGCUUCACUUCGAUGUCGUUGUCCUUGAAGCGCUGCAA-UGAAAAAGACGAAAACCAA---CAAAUCACAAAUA---------- .((((.((((((((((......)))))...((((((....(((......)))..)))))))))))..-))))...............---.............---------- ( -21.30) >DroPer_CAF1 16013 111 + 1 GUUCACGCAGUAGGAUAAUGCCGUCCUUGCGCUUCACCUCGAUGUCGUUGUCCUUGAAGCGCUGCAAAUGGACAAGCGGAAAAGCAAAUAUAAUUUCCAAAUACGAUGCGU-- ....(((((((.((......))((((..((((((((....(((......)))..)))))))).......))))..))(((((...........)))))........)))))-- ( -29.00) >consensus AUUCACGCAGCAGGAUAAUGCCAUCCUUGCGCUUCACCUCGAUGUCGUUGUCCUUGAAGCGCUGCAAAUG_ACAAGCGGAAAAGCA_______UU__CAAA____________ ......((((((((((((((.((((...............)))).)))))))))......)))))................................................ (-19.51 = -18.96 + -0.55)
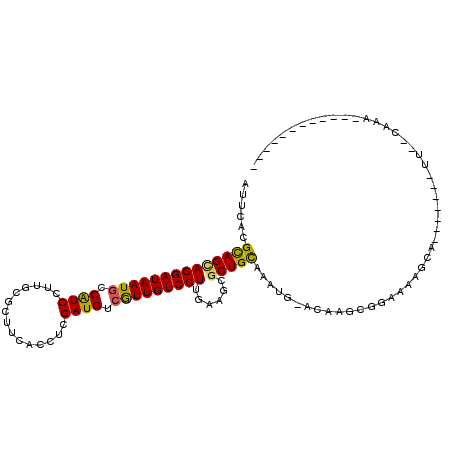
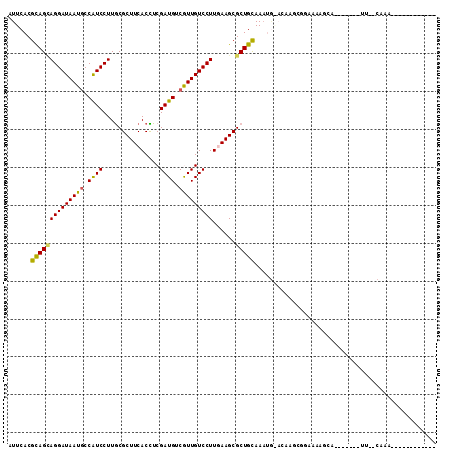
| Location | 15,659,562 – 15,659,652 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 70.22 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -17.94 |
| Energy contribution | -17.72 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.19 |
| Mean z-score | -0.63 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.646785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15659562 90 - 22407834 --A---------UCUGAUGA------------UUUCGUUUUUUCAUUUGCAGCGCUUCAAGGACAACGACAUCGAAGUGAAGCGCAAGGAUGGUAUUAUCCUCCUGCGUGAAU --.---------...((((.------------...))))..(((((..((.((((((((.......((....))...)))))))).((((.((......))))))))))))). ( -24.10) >DroVir_CAF1 37210 81 - 1 AU-------------------------UUCUCUUU----UG---UUUUGCAGCGCUUUAAGGACAAUGACAUCGAAGUGAAGCGCAAGGAUGGCAUUAUCCUGCUGCGUGAAU ..-------------------------........----..---....(((((((((((..((........))....))))))...((((((....)))))))))))...... ( -19.60) >DroPse_CAF1 18302 111 - 1 --ACGCAUCGUAUUUGGAAAUUAUAUUUGCUUUUCCGCUUGUCCAUUUGCAGCGCUUCAAGGACAACGACAUCGAGGUGAAGCGCAAGGACGGCAUUAUCCUACUGCGUGAAC --(((((..(((...(((((...........)))))(((.((((.((....(((((((....((..((....))..)))))))))))))))))).......)))))))).... ( -29.50) >DroGri_CAF1 20613 73 - 1 ---------------------------------UG----U---UUUUUACAGCGCUUUAAGGACAAUGACAUCGAGGUGAAGCGCAAGGAUGGCAUUAUCCUGCUGCGUGAAU ---------------------------------..----.---........((((....((((.((((.((((...(((...)))...)))).)))).))))...)))).... ( -19.90) >DroWil_CAF1 37846 99 - 1 ----------UAUUUGUGAUUUG---UUGGUUUUCGUCUUUUUCA-UUGCAGCGCUUCAAGGACAACGACAUCGAAGUGAAGCGUAAGGAUGGCAUUAUCCUGCUACGUGAAU ----------.............---...............((((-(.((.((((((((.......((....))...)))))))).((((((....))))))))...))))). ( -20.80) >DroPer_CAF1 16013 111 - 1 --ACGCAUCGUAUUUGGAAAUUAUAUUUGCUUUUCCGCUUGUCCAUUUGCAGCGCUUCAAGGACAACGACAUCGAGGUGAAGCGCAAGGACGGCAUUAUCCUACUGCGUGAAC --(((((..(((...(((((...........)))))(((.((((.((....(((((((....((..((....))..)))))))))))))))))).......)))))))).... ( -29.50) >consensus ____________UUUG__AA_______UGCUUUUCCGCUUGU_CAUUUGCAGCGCUUCAAGGACAACGACAUCGAAGUGAAGCGCAAGGAUGGCAUUAUCCUGCUGCGUGAAU .............................................(((((.((((((((.......((....))...)))))))).((((((....)))))).....))))). (-17.94 = -17.72 + -0.22)

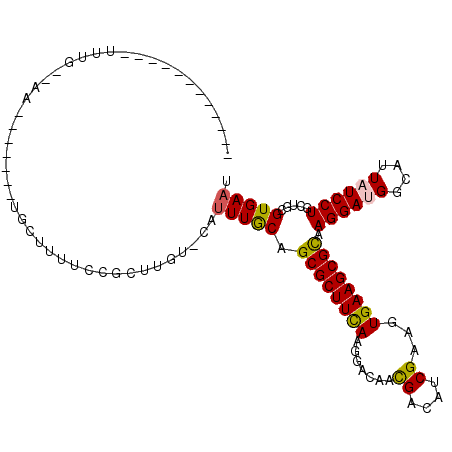
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:16 2006