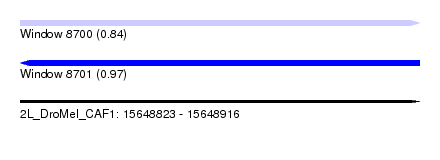
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,648,823 – 15,648,916 |
| Length | 93 |
| Max. P | 0.973940 |

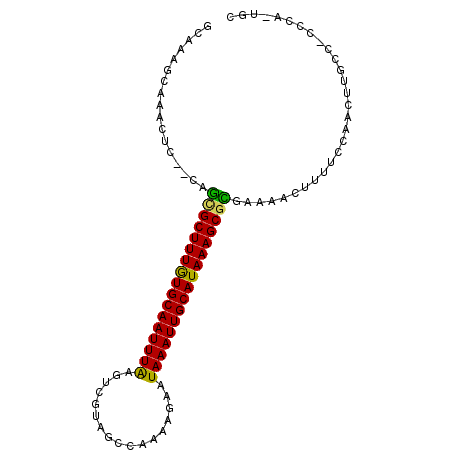
| Location | 15,648,823 – 15,648,916 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 79.27 |
| Mean single sequence MFE | -27.63 |
| Consensus MFE | -16.31 |
| Energy contribution | -15.61 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15648823 93 + 22407834 GAA-UAGG-GGCGAGUUGCAAAACUUUCCGCGCUUUAUGCAAUUUAUUCUUUUCGCUACGACUUAAAUUGCACAAAGCACUG--GAGUUUGCUUGGC ...-...(-((((((((....))).(((((.(((((.(((((((((.((..........))..))))))))).))))).).)--))).))))))... ( -23.90) >DroPse_CAF1 7261 95 + 1 GCA-GGCG-GGAAAGUUGGGCAAGUUUUCCCGCUUUAUGCAAUUUAUUCUCUUGCUCAAGUCUUAAAUUGCACAAAGCGGAGAAGAGUUUGCUUUAC ...-....-.....((..((((((((((((((((((.(((((((((....(((....)))...))))))))).))))))).))))).))))))..)) ( -33.50) >DroSim_CAF1 5208 92 + 1 GAA-UGGG-GGCAAGUUGCAAAACUUUCCGCGCUUUAUGCAAUUUAU-CUUUUCGCUACGACUUAAAUUGCACAAAGCACUG--GAGUUUGCUUGGC ...-...(-((((((((....))).(((((.(((((.(((((((((.-....(((...)))..))))))))).))))).).)--))).))))))... ( -24.40) >DroEre_CAF1 12976 93 + 1 GAA-UGGG-GGCAAGUUUCAAAACUUUUCGCGCUUUAUGCAAUUUAUUCUUUUCGCUACGACCCAAAUUGCACAAAGCUCUG--GAGUUUGCUUGGC ...-....-(.(((((....(((((((..(.(((((.((((((((..((..........))...)))))))).))))).).)--))))))))))).) ( -21.10) >DroAna_CAF1 9398 94 + 1 GCUGUGGGCGGAGUGUUGGAAAAGUUUCCCCGCUUUAUGCAAUUUAUUCUCUCGGAAAGGACUUAAAUUGCAUAAAGCGAAG--GAGUUUCU-UUGC .......((((((....((((....)))).((((((((((((((((.(((........)))..))))))))))))))))...--......))-)))) ( -29.40) >DroPer_CAF1 5043 95 + 1 GCA-GGCG-GGAAAGUUGGGCAAGUUUUCCCGCUUUAUGCAAUUUAUUCUCUUGCUCAAGUCUUAAAUUGCACAAAGCGGAGAAGAGUUUGCUUUAC ...-....-.....((..((((((((((((((((((.(((((((((....(((....)))...))))))))).))))))).))))).))))))..)) ( -33.50) >consensus GAA_UGGG_GGAAAGUUGCAAAACUUUCCCCGCUUUAUGCAAUUUAUUCUCUUCGCUACGACUUAAAUUGCACAAAGCGCAG__GAGUUUGCUUGGC ...........(((((.((..........))(((((.(((((((((.................))))))))).)))))............))))).. (-16.31 = -15.61 + -0.69)

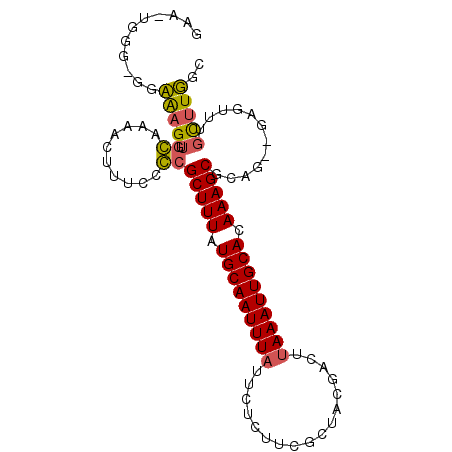
| Location | 15,648,823 – 15,648,916 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 79.27 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -21.86 |
| Energy contribution | -21.03 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15648823 93 - 22407834 GCCAAGCAAACUC--CAGUGCUUUGUGCAAUUUAAGUCGUAGCGAAAAGAAUAAAUUGCAUAAAGCGCGGAAAGUUUUGCAACUCGCC-CCUA-UUC .....(((((.((--(.(((((((((((((((((..((..........)).))))))))))))))))))))....)))))........-....-... ( -30.10) >DroPse_CAF1 7261 95 - 1 GUAAAGCAAACUCUUCUCCGCUUUGUGCAAUUUAAGACUUGAGCAAGAGAAUAAAUUGCAUAAAGCGGGAAAACUUGCCCAACUUUCC-CGCC-UGC .....((((....(((.(((((((((((((((((...(((......)))..))))))))))))))))))))...))))..........-....-... ( -30.00) >DroSim_CAF1 5208 92 - 1 GCCAAGCAAACUC--CAGUGCUUUGUGCAAUUUAAGUCGUAGCGAAAAG-AUAAAUUGCAUAAAGCGCGGAAAGUUUUGCAACUUGCC-CCCA-UUC .....(((((.((--(.(((((((((((((((((..(((...)))....-.))))))))))))))))))))....)))))........-....-... ( -30.20) >DroEre_CAF1 12976 93 - 1 GCCAAGCAAACUC--CAGAGCUUUGUGCAAUUUGGGUCGUAGCGAAAAGAAUAAAUUGCAUAAAGCGCGAAAAGUUUUGAAACUUGCC-CCCA-UUC .....((((....--(((((((((((((((((((..((..........)).)))))))))))))))((.....))))))....)))).-....-... ( -23.50) >DroAna_CAF1 9398 94 - 1 GCAA-AGAAACUC--CUUCGCUUUAUGCAAUUUAAGUCCUUUCCGAGAGAAUAAAUUGCAUAAAGCGGGGAAACUUUUCCAACACUCCGCCCACAGC ((..-........--..(((((((((((((((((..(((((...))).)).)))))))))))))))))((((....))))........))....... ( -27.40) >DroPer_CAF1 5043 95 - 1 GUAAAGCAAACUCUUCUCCGCUUUGUGCAAUUUAAGACUUGAGCAAGAGAAUAAAUUGCAUAAAGCGGGAAAACUUGCCCAACUUUCC-CGCC-UGC .....((((....(((.(((((((((((((((((...(((......)))..))))))))))))))))))))...))))..........-....-... ( -30.00) >consensus GCAAAGCAAACUC__CAGCGCUUUGUGCAAUUUAAGUCGUAGCCAAAAGAAUAAAUUGCAUAAAGCGCGAAAACUUUUCCAACUUGCC_CCCA_UGC .................(((((((((((((((((.................)))))))))))))))))............................. (-21.86 = -21.03 + -0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:15 2006