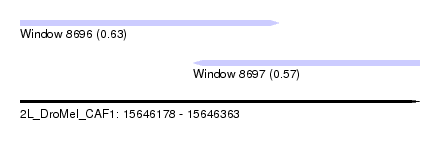
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,646,178 – 15,646,363 |
| Length | 185 |
| Max. P | 0.628757 |

| Location | 15,646,178 – 15,646,298 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.60 |
| Mean single sequence MFE | -44.73 |
| Consensus MFE | -29.37 |
| Energy contribution | -30.63 |
| Covariance contribution | 1.26 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
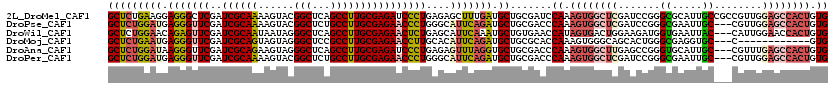
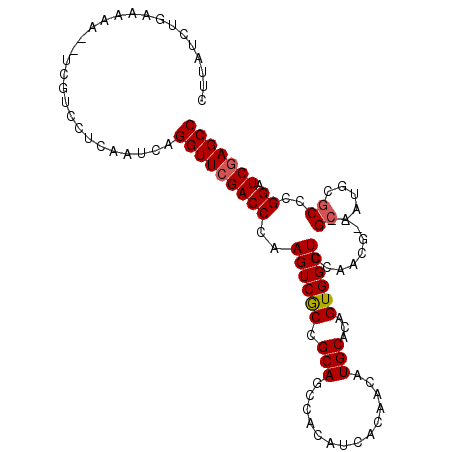
>2L_DroMel_CAF1 15646178 120 + 22407834 GCUCUGAAGGAGGGCUCGAUCGCAAAAGUACGGCUCAGCCUUGCGAGAUCCCUGAGAGCUUUGAUGCUGCGAUCCAAAGUGGCUCGAUCCGGGCGCAUUGCCGCCGUUGGAGCCACUGUG (((((.....)))))..((((((....(((.(((...))).)))............(((......)))))))))((.((((((((.(....((((......))))..).)))))))).)) ( -44.00) >DroPse_CAF1 4769 117 + 1 GCUCUGGAUGAGGGUUCGAUCGCAAAAGUACGGCUCUGCCUUGCGAGAACCCUGGGCAUUCAGAUGCUGCGACCCAAAGUGGCUCGAUCCGGGCGAAUUGC---CGUUGGAGCCACUGUG (((((((((((((((((..((((((......(((...))))))))))))))))...)))))))).)).......((.((((((((.(....(((.....))---)..).)))))))).)) ( -48.50) >DroWil_CAF1 20313 117 + 1 GCUCUGGAACAGAGUUCGAUCGCAAUAAUAGGGCUCAGCCUUGCGAGAACUCUGAGCAUUCAAAUGCUGUGAACCAUAGUGACUGGAAGAUGGUGAAUUAC---CAUUGGAACCACUGUG (((.(((..((((((((..(((((......((((...)))))))))))))))))(((((....))))).....))).)))...(((..(((((((...)))---))))....)))..... ( -37.70) >DroMoj_CAF1 2981 105 + 1 GCUCUGAAUGAGGGUUCGAUCGCAGUAGUAGGGCUCCGCCUUGCGAGAACCUUGCACAUUCAGAUGCUGCGCACCAAAGUGGGCAGCACUGGGCGAGGUGC---C------------GUG .(((.....)))(((....((((.((.((((((.(((((...))).)).))))))))..((((.((((((.(((....))).))))))))))))))...))---)------------... ( -44.80) >DroAna_CAF1 6997 117 + 1 GCUCUGGAUAAGGGUUCGAUCGCAGAAGUAGGGCUCAGCCUUGCGAGAUCCCUGAGAGUUUAGGUGCUGCGACCCAAAGUGGCUUGAGCCGGGUGCAUUGC---CGUUUGAGCCACUGUG (((((.....)))))....((((((..(((((((...)))))))......(((((....)))))..))))))..((.(((((((..(((..(((.....))---))))..))))))).)) ( -44.90) >DroPer_CAF1 2551 117 + 1 GCUCUGGAUGAGGGUUCGAUCGCAAAAGUACGGCUCUGCCUUGCGAGAACCCUGGGCAUUCAGAUGCUGCGACCCAAAGUGGCUCGAUCCGGGCGAAUUGC---CGUUGGAGCCACUGUG (((((((((((((((((..((((((......(((...))))))))))))))))...)))))))).)).......((.((((((((.(....(((.....))---)..).)))))))).)) ( -48.50) >consensus GCUCUGGAUGAGGGUUCGAUCGCAAAAGUACGGCUCAGCCUUGCGAGAACCCUGAGCAUUCAGAUGCUGCGACCCAAAGUGGCUCGAUCCGGGCGAAUUGC___CGUUGGAGCCACUGUG (((((((((.(((((((..((((((......(((...))))))))))))))))....))))))).)).......((.((((((((.......((.....))........)))))))).)) (-29.37 = -30.63 + 1.26)
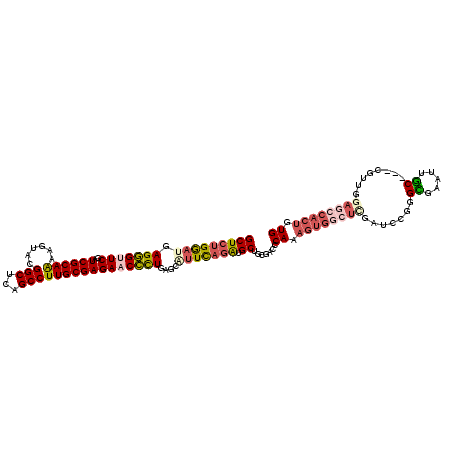
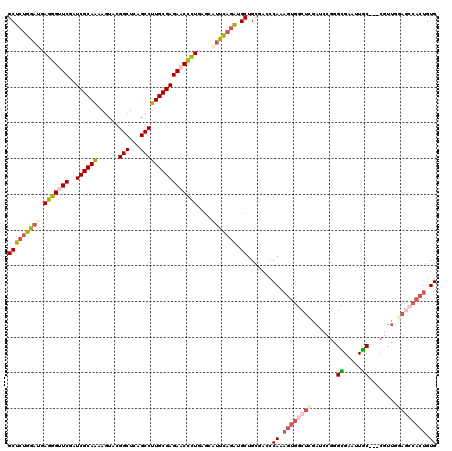
| Location | 15,646,258 – 15,646,363 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.51 |
| Mean single sequence MFE | -26.19 |
| Consensus MFE | -17.51 |
| Energy contribution | -17.71 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15646258 105 - 22407834 CUUAUACGGAAAACCUCUUCCUCAAUCAGGUUCGACCCAAGUCGCCGCAGCCACAUCACAACAUGCACAGUGGCUCCAACGGCGGCAAUGCGCCCGGAUCGAGCC .......((((......)))).......(((((((.((.....((((.((((((...............))))))....))))(((.....))).)).))))))) ( -35.06) >DroPse_CAF1 4849 98 - 1 CUAAUCUUUGAU----UUCCUUCCCUCAGGUUCGACCAAAGUCGCCGCAGCCACAUCACAACAUGCACAGUGGCUCCAACG---GCAAUUCGCCCGGAUCGAGCC ............----............(((((((((.........(.((((((...............)))))).)...(---((.....))).)).))))))) ( -25.16) >DroEre_CAF1 10207 102 - 1 CUUAUGUGAAAUAUUUUGUCCCGCAUCAGGUUCGACCCAAGUCGCCGCAGCCACAUCACAACAUGCACAGUGGCUCCAACG---GCAAUGCGCCCGGAUCGAGCC ...(((((..((.....))..)))))..(((((((.((..((.((((.((((((...............))))))....))---))...))....)).))))))) ( -27.76) >DroYak_CAF1 11365 102 - 1 CUUAUACGAAAAAUCUCGUCUUGAAUCAGGUUCGACCUAAGUCGCCGCAGCCACAUCACAACAUGCACAGUGGCUCCAACG---GCAAUGCGCCAGGAUCGAGCC .(((.((((......))))..)))....((((((((((..((.((((.((((((...............))))))....))---))...))...))).))))))) ( -29.56) >DroAna_CAF1 7077 97 - 1 UUUAUC-----AACUCCGAUCUCAUACAGGUUCGACCCAAGUCACCGCAACCACACCACAACAUGCACAGUGGCUCAAACG---GCAAUGCACCCGGCUCAAGCC ......-----.................((((.((.((.((((((.(((..............)))...)))))).....(---(........)))).)).)))) ( -14.44) >DroPer_CAF1 2631 98 - 1 CUAAUCUUUGAU----UUCCUUUCCUCAGGUUCGACCAAAGUCGCCGCAGCCACAUCACAACAUGCACAGUGGCUCCAACG---GCAAUUCGCCCGGAUCGAGCC ............----............(((((((((.........(.((((((...............)))))).)...(---((.....))).)).))))))) ( -25.16) >consensus CUUAUCUGAAAAA__UCGUCCUCAAUCAGGUUCGACCCAAGUCGCCGCAGCCACAUCACAACAUGCACAGUGGCUCCAACG___GCAAUGCGCCCGGAUCGAGCC ............................(((((((((..((((((.(((..............)))...)))))).........((.....))..)).))))))) (-17.51 = -17.71 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:11 2006