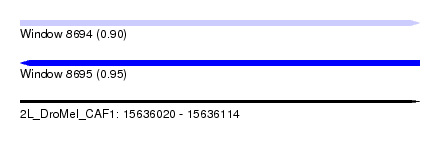
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,636,020 – 15,636,114 |
| Length | 94 |
| Max. P | 0.948421 |

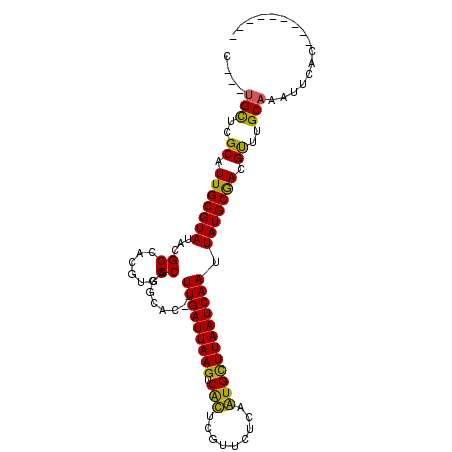
| Location | 15,636,020 – 15,636,114 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.93 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -21.27 |
| Energy contribution | -20.72 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15636020 94 + 22407834 ---------GUGAAUUUGCAAACGUCGCAUAAUUGAUUAAGCACGGAGAACGAGCGACUUAAUCAACGGUGCGAGCAACGUGGCGUAUACGCAAUGCGAGCA---G ---------.......(((.....((((((..((((((((((.((.....)).))...))))))))..))))))....(((.(((....)))...))).)))---. ( -28.40) >DroVir_CAF1 193199 101 + 1 GGCAAGCUUGUGAAUUUGCAAGCAUUGCAUAAUUGAUUAAACAUUGAAAACGAAUGACUUAAUCAA--GUGCGUGCCACGUGGCGUAUACGCAAUGCGAGCA---A .((..((((((......)))))).((((((..((((((((.((((.......))))..))))))))--.((((((((....)))....))))))))))))).---. ( -31.90) >DroGri_CAF1 153906 104 + 1 GACGGACUGGCAAAUUUGCAAACGUCGCAUAAUUGAUUAAACAUUGAAAUCGAAUGACUUAAUCAA--GUGCGUGCCACGUGGCGUAUACGCAAUGCGAACAACAA ((((.....(((....)))...))))((((..((((((((.((((.......))))..))))))))--.((((((((....)))....)))))))))......... ( -27.80) >DroSim_CAF1 136366 94 + 1 ---------GUGAAUUUGCAAACGUCGCAUAAUUGAUUAAGCACGGAGAACGAGCGACUUAAUCAACGGUGCGAGCAACGUGGCGUAUACGCAAUGCGAGCA---G ---------.......(((.....((((((..((((((((((.((.....)).))...))))))))..))))))....(((.(((....)))...))).)))---. ( -28.40) >DroEre_CAF1 140047 94 + 1 ---------GUGAAUUUGCAAACGUCGCAUAAUUGAUUAAGCACGGCGAACGAGCGACUUAAUCAACGGUGCGAGCAACGUGGCGUAUACGCAAUGCGAGCA---G ---------.......(((.....((((((..(((((((((....((......))..)))))))))..))))))....(((.(((....)))...))).)))---. ( -29.20) >DroMoj_CAF1 189834 90 + 1 -----------GCAUUUGCAAGCGUUGCAUAAUUGAUUAAACAUUCAAAUCGAAUGACUUAAUCAA--GUGUGUGCCACGUGGCGUAUACGCAAUGCGAGCG---C -----------......((..(((((((....((((((((.(((((.....)))))..))))))))--(((((((((....))))))))))))))))..)).---. ( -34.90) >consensus _________GUGAAUUUGCAAACGUCGCAUAAUUGAUUAAACACGGAAAACGAACGACUUAAUCAA__GUGCGAGCAACGUGGCGUAUACGCAAUGCGAGCA___G ..............((((((.(((((((((..((((((((..................))))))))..)))))....)))).(((....)))..))))))...... (-21.27 = -20.72 + -0.55)

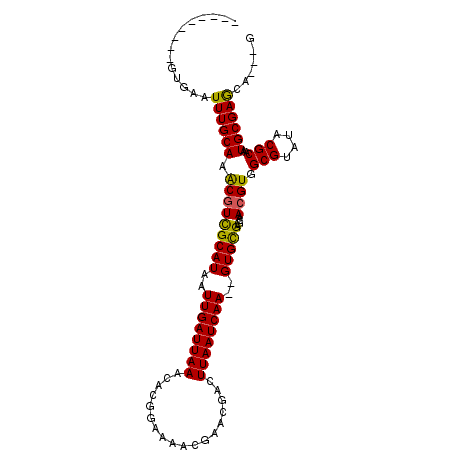
| Location | 15,636,020 – 15,636,114 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.93 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -23.13 |
| Energy contribution | -21.72 |
| Covariance contribution | -1.41 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15636020 94 - 22407834 C---UGCUCGCAUUGCGUAUACGCCACGUUGCUCGCACCGUUGAUUAAGUCGCUCGUUCUCCGUGCUUAAUCAAUUAUGCGACGUUUGCAAAUUCAC--------- .---(((..(((..(((....))).....)))(((((..((((((((((.(((.........)))))))))))))..))))).....))).......--------- ( -27.40) >DroVir_CAF1 193199 101 - 1 U---UGCUCGCAUUGCGUAUACGCCACGUGGCACGCAC--UUGAUUAAGUCAUUCGUUUUCAAUGUUUAAUCAAUUAUGCAAUGCUUGCAAAUUCACAAGCUUGCC (---(((..((((((((((...(((....)))......--(((((((((.((((.......))))))))))))).))))))))))..))))............... ( -31.30) >DroGri_CAF1 153906 104 - 1 UUGUUGUUCGCAUUGCGUAUACGCCACGUGGCACGCAC--UUGAUUAAGUCAUUCGAUUUCAAUGUUUAAUCAAUUAUGCGACGUUUGCAAAUUUGCCAGUCCGUC .........(((..((((....(((....))).((((.--(((((((((.((((.(....))))))))))))))...)))))))).)))................. ( -26.00) >DroSim_CAF1 136366 94 - 1 C---UGCUCGCAUUGCGUAUACGCCACGUUGCUCGCACCGUUGAUUAAGUCGCUCGUUCUCCGUGCUUAAUCAAUUAUGCGACGUUUGCAAAUUCAC--------- .---(((..(((..(((....))).....)))(((((..((((((((((.(((.........)))))))))))))..))))).....))).......--------- ( -27.40) >DroEre_CAF1 140047 94 - 1 C---UGCUCGCAUUGCGUAUACGCCACGUUGCUCGCACCGUUGAUUAAGUCGCUCGUUCGCCGUGCUUAAUCAAUUAUGCGACGUUUGCAAAUUCAC--------- .---(((..(((..(((....))).....)))(((((..(((((((((((.((......))...)))))))))))..))))).....))).......--------- ( -27.40) >DroMoj_CAF1 189834 90 - 1 G---CGCUCGCAUUGCGUAUACGCCACGUGGCACACAC--UUGAUUAAGUCAUUCGAUUUGAAUGUUUAAUCAAUUAUGCAACGCUUGCAAAUGC----------- .---.((..((.(((((((...(((....)))......--(((((((((.(((((.....)))))))))))))).))))))).))..))......----------- ( -29.40) >consensus C___UGCUCGCAUUGCGUAUACGCCACGUGGCACGCAC__UUGAUUAAGUCACUCGUUCUCAAUGCUUAAUCAAUUAUGCGACGUUUGCAAAUUCAC_________ ....(((..((.(((((((...((......))........(((((((((.(((.........)))))))))))).))))))).))..)))................ (-23.13 = -21.72 + -1.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:09 2006