| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,618,569 – 15,618,684 |
| Length | 115 |
| Max. P | 0.563374 |

| Location | 15,618,569 – 15,618,684 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.84 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -17.60 |
| Energy contribution | -16.75 |
| Covariance contribution | -0.85 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15618569 115 - 22407834 UCAGUAUACUGUGCGGGUUUUUAUUAUGUUUGCAUUCGCUGCGGGCCUUUUAUGUGCUCUUUCUUUUAAGGCUUAUACGGUAAGCAAAAAAC-AAUAGAAAUAUUAUUUCAACUCU ...((.(((((((..((((((..........(((.....)))(((((......).))))........))))))..))))))).)).......-((((....))))........... ( -22.80) >DroSim_CAF1 118853 112 - 1 UCAGUAUACUGUGCGGGUUUUUAUUAUGUUUGCAUUCGCUGCGGGCCUUUUAUGUGCGCUUUCUUUUAAGGCUUAUACGGUAAGCAAAAAAC-AAUAAAAAUAA---UUAAACUCU .(((....))).....(((((((((...(((((..(((.((..((((((....(........)....))))))..)))))...)))))....-)))))))))..---......... ( -21.80) >DroEre_CAF1 120924 110 - 1 UCAGUAUACUGUGCGGGUUUUUAUUAUGUUUGCAUUCGCUGCGGGCCUUUUAUGUGCGCUUUCUUUUAAGGCUUAUACGGUGAGCAAAAAAA-AAUAAU---AAAAUAUCAACU-- .(((....))).(..((((((((((((.(((((..((((((.(((((((....(........)....)))))))...)))))))))))....-.)))))---)))).)))..).-- ( -25.30) >DroWil_CAF1 178274 111 - 1 ACAAUACACAGUUAGAGUUUUCAUAAUGUUUGCAUUUGCUGCUGGUCUACUGUGUGCCCUAUCCUUCAAGGCUUAUACGGUGGGUCGAUUAC-AUAAUGCAUUA---UUG-ACUCC ..............(((((...(((((((..(((.....)))((..((((((((((.(((........)))..))))))))))..)).....-.....))))))---).)-)))). ( -28.80) >DroYak_CAF1 123825 113 - 1 UCAGUAUACUGUACGGGUUUUUAUUAUGUUUGCAUUCGCUGCGGGCCUUUUAUGCGCUCUUUCUUUCAAAGCUUAUACGGUGAGCAAAAAAAAGAUAAU---AUAGUCUGAACUAC ...((.(((((((..((((((..........(((.....)))(((((......).))))........))))))..))))))).)).......((((...---...))))....... ( -23.50) >DroAna_CAF1 358390 113 - 1 ACAGUACACCGUGCGGGUCUUUAUUAUGUUUGCAUUUGCGGCAGGACUUUUGUGUGCCUUAUCCUUUAAGGCCUAUACGGUGGGUAAUUGAAAAGUUAAAUGAA---UUGAAGUGU .((((.(((((((..((((((..........((....))((((.(.(....)).)))).........))))))..)))))))..(((((....))))).....)---)))...... ( -32.10) >consensus UCAGUAUACUGUGCGGGUUUUUAUUAUGUUUGCAUUCGCUGCGGGCCUUUUAUGUGCCCUUUCUUUUAAGGCUUAUACGGUGAGCAAAAAAA_AAUAAAAAUAA___UUGAACUCU ...((.(((((((..((((((..........(((.....)))(((((......).))))........))))))..))))))).))............................... (-17.60 = -16.75 + -0.85)

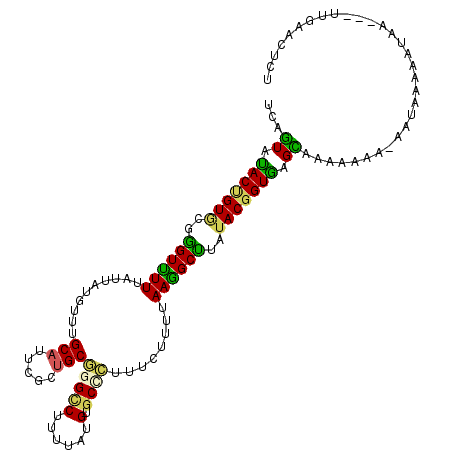

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:59 2006