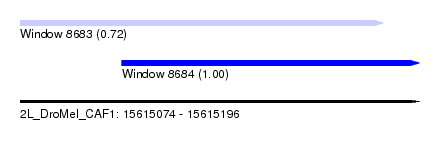
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,615,074 – 15,615,196 |
| Length | 122 |
| Max. P | 0.999747 |

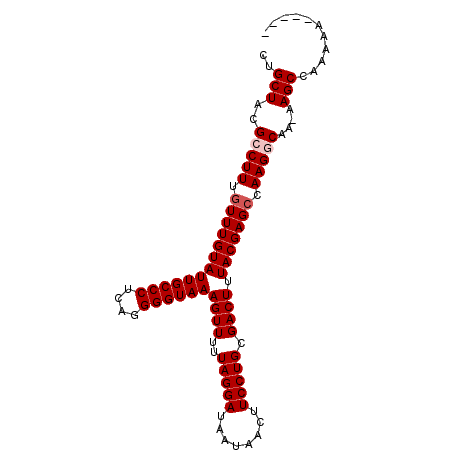
| Location | 15,615,074 – 15,615,185 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.79 |
| Mean single sequence MFE | -33.35 |
| Consensus MFE | -28.63 |
| Energy contribution | -29.30 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15615074 111 + 22407834 UAGUCAGACAUGAAUGCUG---------CGCUUUGUUGCCCUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAGGCA ...(((....)))..((.(---------.((......)).).))...(((((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).))))). ( -33.70) >DroVir_CAF1 164966 110 + 1 UAGUCAGACAUGAAUGCUG---------CGCUUUGUUGCUGUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGGCAAG-CA ...(((....))).(((((---------(((.........))))...(((((...........(((...)))((((((((..(((((........))))).))))))))))))).))-)) ( -30.90) >DroSec_CAF1 121911 111 + 1 UAGUCAGACAUGAAUGCUG---------CGCUUUGUUGCCCUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGUCUUUACGAGCCAAGGCA ...(((....)))..((.(---------.((......)).).))...(((((.(((((((((((((....))))))......(((((........)))))......))))))).))))). ( -32.10) >DroWil_CAF1 173741 120 + 1 UAGUCAGACAUGAAUGCUGUUGCUGCAGUGCUUUGUUGCUCUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAGGCA (((.((((((.((..(((((....)))))..))...)).))))))).(((((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).))))). ( -41.50) >DroYak_CAF1 120202 111 + 1 UAGUCAGACAUGAAUGCUG---------AGCUUUGUUGCCCUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAGGCA ...((((.((....)))))---------)((......))........(((((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).))))). ( -34.00) >DroMoj_CAF1 161563 110 + 1 UAGUCAGACAUGAAUGCUG---------CGCUUUGUUGCUGUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAG-CA (((((((..(..((.((..---------.))))..)..))).)))).((.((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).)))-). ( -27.90) >consensus UAGUCAGACAUGAAUGCUG_________CGCUUUGUUGCCCUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAGGCA (((((((.((....)))))..........((......))...)))).(((((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).))))). (-28.63 = -29.30 + 0.67)

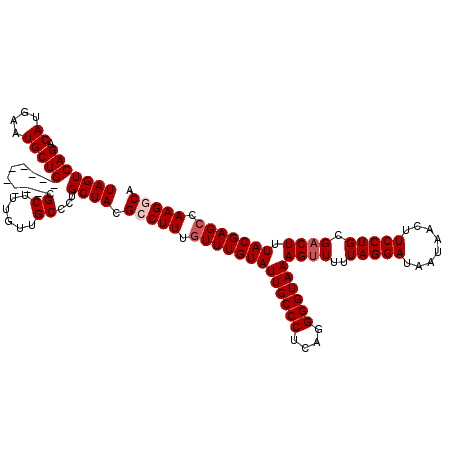
| Location | 15,615,105 – 15,615,196 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 93.10 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -26.85 |
| Energy contribution | -27.35 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.99 |
| SVM RNA-class probability | 0.999747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15615105 91 + 22407834 CUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAGGCAA-AAGCCAAAAA----- ..(((..(((((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).)))))..-.)))......----- ( -30.80) >DroVir_CAF1 164997 95 + 1 GUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGGCAAG-CAA-AAGCCAAAACCCAGC .......((.(((((((((....(((...)))((((((((..(((((........))))).))))))))...)))))-)))-).))........... ( -27.50) >DroPse_CAF1 177218 91 + 1 CUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAGGCAA-AAGCCGAAUU----- ..(((..(((((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).)))))..-.)))......----- ( -30.80) >DroWil_CAF1 173781 94 + 1 CUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAGGCAAAAAGCCAAAAA---AC ..(((..(((((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).)))))....)))......---.. ( -30.10) >DroMoj_CAF1 161594 95 + 1 GUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAG-CAA-AAGCCAAAACCCAGC .......((.((((((((..((.(((...)))((((((((..(((((........))))).))))))))))..))))-)))-).))........... ( -25.50) >DroPer_CAF1 176423 91 + 1 CUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAGGCAA-AAGCCGAAUU----- ..(((..(((((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).)))))..-.)))......----- ( -30.80) >consensus CUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAGGCAA_AAGCCAAAAA_____ ..(((..(((((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).)))))....)))........... (-26.85 = -27.35 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:58 2006