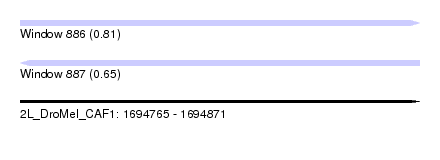
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,694,765 – 1,694,871 |
| Length | 106 |
| Max. P | 0.809560 |

| Location | 1,694,765 – 1,694,871 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.88 |
| Mean single sequence MFE | -54.27 |
| Consensus MFE | -30.67 |
| Energy contribution | -29.53 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
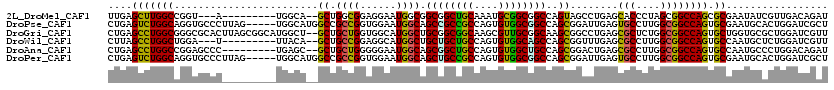
>2L_DroMel_CAF1 1694765 106 + 22407834 UUGAGCUUGGCCGGU---A---------UGGCA--GCUGGCGGAGGAAUGGCGGCGGCUGCAAAUGCGGCGGCCAGUAGCCUGAGCACCCUAGCGGCCAGCGCGAAUAUCGUUGACAGAU ....(.(..((.(((---(---------(.((.--((((((..(((....(((((.(((((....))))).))).))..)))..((......)).))))))))..)))))))..)).... ( -43.00) >DroPse_CAF1 8301 115 + 1 CUGAGUCUGGCAGGUGCCCUUAG-----UGGCAUGGCCGCCGGUGGAAUGGCAGCCGCCGCCAGUGUGGCGGCCAGCGGAUUGAGUGCCUUGGCGGCCAGUGCGAAUGCACUGGAUCGCU ((.((((((((..(((((.....-----.))))).)))((((......)))).((((((((....))))))))....))))).))......((((((((((((....))))))).))))) ( -57.50) >DroGri_CAF1 5523 118 + 1 CUGAGCCUGGCGGGCGCACUUAGCGGCAUGGCU--GCUGCUGGUGGCAUGGCUGCGGCGGCAAGCGUUGCGGCAAGCGGCCUGAGCGCUCUGGCGGCCAGUGCUGGUGCGCUGGAUCGUU ..((((....(((((.((((.(((((((....)--))))))))))((...(((((((((.....)))))))))..)).)))))...)))).((((((((((((....))))))).))))) ( -62.40) >DroWil_CAF1 6863 106 + 1 CUUAGCCUGGCUGGA---U---------UUACA--GCUGCCGGAGGCAUGGCUGCUGCUGCCAGUGUGGCAGCCAGCGGUUUGAGCGCCUUGGCGGCCAGUGCCAAUGCUCUGGAUCGUU ....(((.(((((..---.---------...))--)))((((.((((..(((((((((((((.....))))).)))))))).....)))))))))))(((.((....)).)))....... ( -49.60) >DroAna_CAF1 4511 109 + 1 CUGAGCCUGGCCGGAGCCC---------UGAGC--GCUGCUGGGGGAAUGGCAGCGGCUGCCAGUGUGGCUGCCAGCGGACUGAGCGCCUUGGCGGCCAGUGCCAAUGCCCUGGACAGAU ....(((.(((....))).---------.).))--.((((..(((.(.(((((..(((((((((...(((.(((((....))).))))))))))))))..))))).).)))..).))).. ( -53.80) >DroPer_CAF1 4970 115 + 1 CUGAGUCUGGCAGGUGCCCUUAG-----UGGCAUGGCCGCCGGUGGAAUGGCAGCUGCCGCCAGUGUGGCGGCCAGCGGAUUGAGUGCCUUGGCGGCCAGUGCGAAUGCACUGGAUCGCU ((.((((((((..(((((.....-----.))))).)))((((......)))).(((((((((.....))))).))))))))).))......((((((((((((....))))))).))))) ( -59.30) >consensus CUGAGCCUGGCAGGUGCCC_________UGGCA__GCUGCCGGUGGAAUGGCAGCGGCUGCCAGUGUGGCGGCCAGCGGACUGAGCGCCUUGGCGGCCAGUGCGAAUGCACUGGAUCGAU ....(((((((........................((.((((......)))).((((((((....))))))))..))........(((....)))))))).))................. (-30.67 = -29.53 + -1.13)
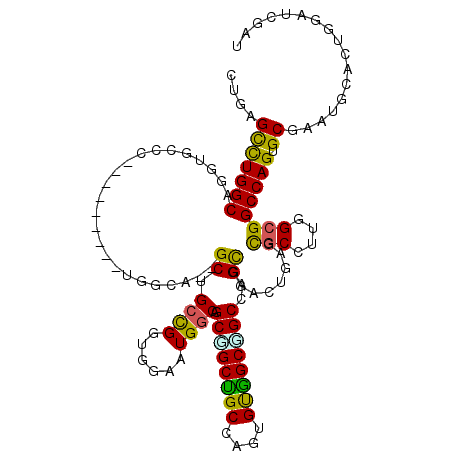
| Location | 1,694,765 – 1,694,871 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.88 |
| Mean single sequence MFE | -45.70 |
| Consensus MFE | -19.88 |
| Energy contribution | -20.93 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1694765 106 - 22407834 AUCUGUCAACGAUAUUCGCGCUGGCCGCUAGGGUGCUCAGGCUACUGGCCGCCGCAUUUGCAGCCGCCGCCAUUCCUCCGCCAGC--UGCCA---------U---ACCGGCCAAGCUCAA .................(.((((((((.((.((((((..(((...((((.((.((.......)).)).)))).......))))))--.))).---------)---).))))).))).).. ( -34.80) >DroPse_CAF1 8301 115 - 1 AGCGAUCCAGUGCAUUCGCACUGGCCGCCAAGGCACUCAAUCCGCUGGCCGCCACACUGGCGGCGGCUGCCAUUCCACCGGCGGCCAUGCCA-----CUAAGGGCACCUGCCAGACUCAG .(((..(((((((....))))))).)))...((((............((((((.....))))))(((((((........))))))).)))).-----.....(((....)))........ ( -55.20) >DroGri_CAF1 5523 118 - 1 AACGAUCCAGCGCACCAGCACUGGCCGCCAGAGCGCUCAGGCCGCUUGCCGCAACGCUUGCCGCCGCAGCCAUGCCACCAGCAGC--AGCCAUGCCGCUAAGUGCGCCCGCCAGGCUCAG ...((.((.((((((..(((.((((.((..((....)).(((.((..((......))..)).))))).)))))))....(((.((--(....))).)))..))))))......)).)).. ( -41.30) >DroWil_CAF1 6863 106 - 1 AACGAUCCAGAGCAUUGGCACUGGCCGCCAAGGCGCUCAAACCGCUGGCUGCCACACUGGCAGCAGCAGCCAUGCCUCCGGCAGC--UGUAA---------A---UCCAGCCAGGCUAAG .........((((...(((....)))((....)))))).....((((.(((((.....)))))))))((((.((((...))))((--((...---------.---..))))..))))... ( -44.90) >DroAna_CAF1 4511 109 - 1 AUCUGUCCAGGGCAUUGGCACUGGCCGCCAAGGCGCUCAGUCCGCUGGCAGCCACACUGGCAGCCGCUGCCAUUCCCCCAGCAGC--GCUCA---------GGGCUCCGGCCAGGCUCAG ..((((...(((...(((((.((((.((((.(((((.(((....))))).)))....)))).)))).)))))...)))..)))).--(((..---------.(((....))).))).... ( -47.10) >DroPer_CAF1 4970 115 - 1 AGCGAUCCAGUGCAUUCGCACUGGCCGCCAAGGCACUCAAUCCGCUGGCCGCCACACUGGCGGCAGCUGCCAUUCCACCGGCGGCCAUGCCA-----CUAAGGGCACCUGCCAGACUCAG .(((..(((((((....))))))).)))...((((............((((((.....)))))).((((((........))))))..)))).-----.....(((....)))........ ( -50.90) >consensus AACGAUCCAGGGCAUUCGCACUGGCCGCCAAGGCGCUCAAUCCGCUGGCCGCCACACUGGCAGCCGCUGCCAUUCCACCGGCAGC__UGCCA_________GGGCACCGGCCAGGCUCAG ......((((.((....)).))))..((....))..........(((((.(((.....))).(((((.(((........))).))..)))...................)))))...... (-19.88 = -20.93 + 1.06)

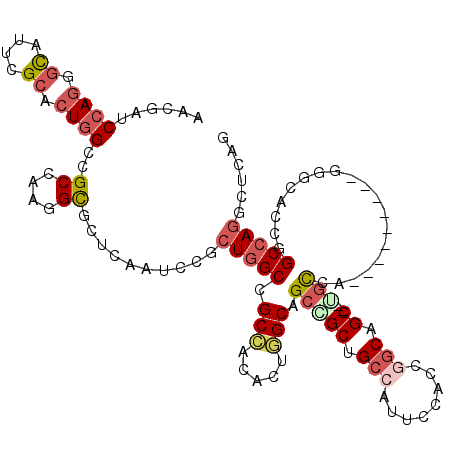
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:04 2006